**Disclaimer:** The two sections *A simple "reproducible research" emacs configuration* and *A stub of replicable article* explain how to set up emacs/org-mode for this MOOC. These are very important sections in the context of this MOOC. **These sections are illustrated in two out of the [three video tutorials of this sequence](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4), and****which you really should follow carefully**. **Otherwise, you may have trouble doing the exercises later on**. Likewise, I strongly encourage you to watch the ["emacs and git" video tutorial available at the same place](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4).
The next section provides information on how to install emacs.
Table of Contents<span class="tag" data-tag-name="TOC"></span>
We provide here only instructions for Debian-based distributions. Feel free to contribute to this document to provide up-to-date information for other distributions (e.g.n redhat, fedora).
Today, the stable versions of the most common distributions provide recent enough versions of emacs and org-mode:
- Debian (stretch) ships with [emacs 25.1](https://packages.debian.org/stretch/emacs25) and [org-mode 9.0.3](https://packages.debian.org/stretch/org-mode)
- Ubuntu (bionic 18.04) ships with [emacs 25.2](https://packages.ubuntu.com/bionic/emacs25) and [org-mode 9.1.6](https://packages.ubuntu.com/bionic/org-mode)
- Ubuntu (artful 17.04) ships with [emacs 25.2](https://packages.ubuntu.com/artful/emacs25) and [org-mode 9.0.9](https://packages.ubuntu.com/artful/org-mode)
If your distribution is older than this, well, it may be a good time for upgrading...
Then make sure you have a sufficiently recent version of emacs.
``` bash
emacs --version 2>&1 | head-n 1
```
``` example
GNU Emacs 25.2.2
```
Likewise, you'll want to check you have a recent version of org-mode:
``` bash
emacs -batch--funcall"org-version" 2>&1 | grep version
```
``` example
Org mode version 9.1.11 (9.1.11-dist @ /usr/share/emacs/25.2/site-lisp/elpa/org-9.1.11/)
```
The version numbers you get will depend on the distribution you are running. **You really want to make sure you do not rely on org-mode 8**, which is now deprecated.
macOS
-----
**Note:** macOS comes with a prehistoric command-line-only version of Emacs located at `/usr/bin/emacs`. It's best to forget about it.
-**Option 1**: Install the `.dmg` file from [Vincent Goulet](http://vgoulet.act.ulaval.ca/): [<https://vigou3.gitlab.io/emacs-modified-macos/>](https://vigou3.gitlab.io/emacs-modified-macos/). It ships with recent versions:
- Emacs 26.1
- Org-mode 9.1.13
- ESS 17.11
If you install this version of Emacs, or in fact any other version of Emacs distributed as a clickable application in a `.dmg` file, you must type the full path to the executable if you want to run Emacs from a terminal. For example, if your clickable application is at `/Applications/Emacs.app`, then the executable is at `/Applications/Emacs.app/Contents/MacOS/Emacs`
-**Option 2**: If you use [Homebrew](https://docs.brew.sh/), do the following:
``` bash
brew update
brew install emacs --with-cocoa
brew linkapps emacs
brew install wget
brew tap dunn/emacs
brew install auctex
brew tap brewsci/science
brew install ess
```
This provides an `emacs` command for use from the command line, plus a clickable application at `Cellar/emacs/26.1_1/Emacs.app` inside your Homebrew directory. If you installed Homebrew at the default location `/usr/local`, then this is `/usr/local/Cellar/emacs/26.1_1/Emacs.app`. If you installed Homebrew on an account with administrator privileges, you can add
``` bash
brew linkapps emacs
```
in order to make Emacs accessible directly from `/Applications`.
Windows
-------
Install the `.exe` file from [Vincent Goulet](http://vgoulet.act.ulaval.ca/): [<https://vigou3.gitlab.io/emacs-modified-windows/>](https://vigou3.gitlab.io/emacs-modified-windows/). It ships with recent versions:
- Emacs 26.1
- Org-mode 9.1.13
- ESS 17.11
### Directory naming conventions
In the following instructions, we refer to your home directory through the (UNIX) `~/` notation. On Windows, your home directory should be something like `C:\Users\yourname`. Therefore, whenever we mention the `~/org/` (resp. the `~/.emacs.d/`) directory this means we are referring to `C:\Users\yourname\org` (resp. `C:\Users\yourname\.emacs.d\`).
### Making R and Python available to the console
When running a command, Windows will look for the command in the directories indicated in the `PATH` environment variable. If none of these directories contains the command, Windows will stop and indicate the command does not exist. To make sure R (which may be in something like `C:/Program Files/R/R-3.5.1/bin/x64/`) and Python (which may be in something like `C:/Program Files/Python/Python37/`) can easily be run from Emacs, you should thus configure the `PATH` variable accordingly.
This requires to go through the "Environment Variable" editor as explained [here](http://sametmax.com/ajouter-un-chemin-a-la-variable-denvironnement-path-sous-windows/).
### Installing and configuring Matplotlib (graphic python library)
Open an DOS console and type the following command:
``` shell
python -m pip install -U matplotlib
```

Then you will want to deactivate interactive plots in matplotlib. To this end, you first need to know where the matplotlib configuration is located. Open a python console the type the following code:
``` python
import matplotlib
matplotlib.matplotlib_fname()
```

Open the `matplotlibrc` file and modify the line starting with `backend` to make it `backend : Agg`.
All platforms: pretty code in HTML export
-----------------------------------------
To have code pretty printing when exporting to HTML, you should install the `htmlize` package, which is done by opening emacs and typing the following command:
``` example
M-x package-install RET htmlize RET # where M-x means pressing the "Esc" key then the "x" key
```
A simple "*reproducible research*" emacs configuration
This section is illustrated in a [video tutorial](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4) (/"Mise en place Emacs/Orgmode"/ in French). Watching it before following the instructions given in this section may help.
Emacs comes with very basic default configuration and it appears like everyone has its own taste. You will for example find [here](https://www.emacswiki.org/emacs/StarterKits) several default Emacs configurations that reflect the preferences of their creators. Likewise the configuration of Org-Mode is incredibly flexible (see for example [the org-mode website](https://orgmode.org/worg/org-configs/index.html) for more references). In the context of this MOOC, we propose you a relatively minimalist one that is rather "*reproducible research*" oriented by adding a few org-mode specific configurations.
Step 0: Backup and download our configuration
---------------------------------------------
The procedure we propose will wipe your already existing custom Emacs configuration if you have one. **You should thus beforehand make a backup** of `~/.emacs` and of `~/.emacs.d/init.el` (if these files exist).
Then download [this archive](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/rr_org_archive.tgz) and uncompress it. It contains the following files and we will refer to them in the following:
``` example
rr_org/init.el
rr_org/journal.org
```
Alternatively, [the files you are looking for are available here](rr_org/).
Step 1: Prepare your journal
----------------------------
Create an `org/` directory in the top of your home:
``` bash
mkdir -p ~/org/
```
Then copy `rr_org/journal.org` file in your `~/org/` directory. This file will be your laboratory notebook and all the notes you will capture with `C-c c` will go automatically go in this file. The first entry of this notebook is populated with [many Emacs shortcuts](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/rr_org/journal.org) that you should give a try.
Step 2: Set up Emacs configuration
----------------------------------
Copy `rr_org/init.el` in your `~/.emacs.d/` directory.
Alternatively, if you do not want to mess with your already existing emacs configuration, you may launch emacs with this specific configuration with the following command: `emacs -q -l rr_org/init.el`.
Step 3: Adapt the configuration to your specific needs if required
Open a new instance of Emacs and open a `foo.org` file. Copy the following lines in this file:
``` example
#+begin_src shell :session foo :results output :exports both
ls -la # or dir under windows
#+end_src
```
Put your cursor inside this code block and execute it with the following command: `C-c C-c` (If you are not familiar with Emacs commands, this one means '`Ctrl + C`' twice)
A `#+RESULTS:` block with the result of the command should appear if it worked.
In the video, we already have demonstrated the main features and shortcuts of emacs/org-mode that will help you maintain a document and benefit from literate programming. The list of features and shortcuts is demonstrated in the [first entry of your labbook](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/rr_org/journal.org).
Step 5: Open and play with your journal:
----------------------------------------
In step 1, you were told to create an journal in `~org/journal.org`. First you probably want to make sure this file is stored in a version control system like git. We leave it up to you to set this up but if you have any trouble, feel free to ask on the FUN forums.
A stub of a replicable article
==============================
This section is illustrated in a [video tutorial](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4) (/"Écrire un article réplicable avec Emacs/Orgmode"/ in French). Watching it before following the instructions given in this section may help.
Remember, you need a working LaTeX and R environment. If you can't open a terminal and run the commands `R`, `pdflatex`, and `python`, you will not be able to generate this document. When being compiled, the article downloads the corresponding LaTeX packages so you also need to have a working `wget` command (alternatively, it uses `curl`). Once downloaded, you may still read the source (<https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/replicable_article/article.org>) and understand how it works though.
Download the following [archive](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/replicable_article.tgz), uncompress it and simply `make` to generate the article. You should then be able to open the [resulting article](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/replicable_article/article.pdf). This is summarized in the following command:
tar zxf replicable_article.tgz; cd replicable_article; make ; evince article.pdf
```
**Possible issues**:
- If the `make` command fails (especially on Mac), it may be because Emacs or something else is not correctly installed. In that case, open the article directly with the following command:
and export it to pdf with the following shortcut: `C-c C-e l o`
- If it still doesn't work and emacs complains about not finding ESS, it may be because you installed ESS in your home instead of system-wide. In that case, try to remove the `-q` in the previous command line to load your personal emacs configuration.
Finally, when you'll be tired of always re-executing all the source code when exporting, just look for the following line in [article.org](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/replicable_article/article.org):
``` example
# #+PROPERTY: header-args :eval never-export
```
If you remove the `# ` in the beginning of the line, it will not be a comment anymore and will indicate org-mode to stop evaluating every chunk of code when exporting.
Emacs tips and tricks
=====================
Cheat-sheets
------------
Learning Emacs and Org-Mode can be difficult as there is an inordinate amount of shortcuts. Many people have thus come up with cheat-sheats. Here is a selection in case it helps:
### Emacs
- [Common and step-by-step Emacs shortcuts for our *reproducible research* configuration](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/rr_org/journal.org)
- [The official GNU emacs refcard](https://www.gnu.org/software/emacs/refcards/pdf/refcard.pdf)
- Two graphical cheat-sheats by Sacha Chua on 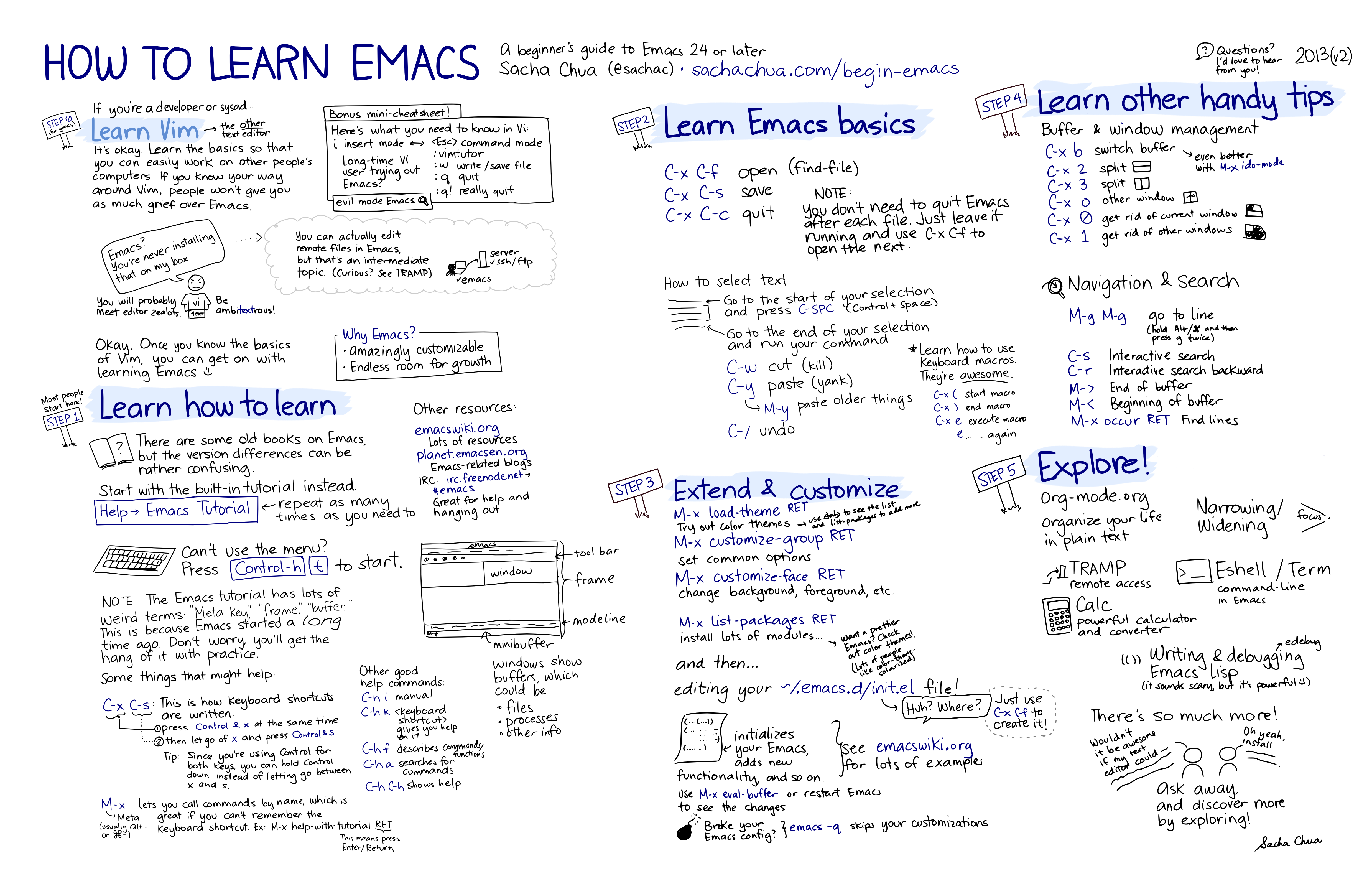 and on .
### Org-mode
- [Common and step-by-step org-mode shortcuts for our *reproducible research* configuration](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/rr_org/journal.org)
- [The official org-mode refcard](https://orgmode.org/worg/orgcard.html)
- [The official description of the org-mode syntax](https://orgmode.org/worg/dev/org-syntax.html) and a [relatively concise description of the org-mode syntax](https://gist.github.com/hoeltgman/3825415).
Video tutorials
---------------
For those of you who prefer video explanations, here is a [Youtube channel with many step by step emacs tutorials](https://www.youtube.com/playlist?list=PL9KxKa8NpFxIcNQa9js7dQQIHc81b0-Xg).
Additional useful emacs packages
--------------------------------
### Company-mode
[Company-mode](http://company-mode.github.io/) is a text completion framework for Emacs. It allows to have smart completion in emacs for the most common languages. If you feel this is needed, you should follow the instructions from the official Web page: [<http://company-mode.github.io/>](http://company-mode.github.io/)
### Magit
[Magit](https://magit.vc/) is an Emacs interface for Git. Its usage is briefly illustrated in the context of this MOOC in a [video tutorial](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4) ("*Utilisation Emacs/git*" in French).
It is very powerful and we use it on a daily basis but you should definitely understand what git does behind the scenes beforehand. If you feel this would be useful for you, you should follow [this visual walk-through](https://magit.vc/screenshots/) or [this really short "crash course"](https://www.emacswiki.org/emacs/Magit). If you installed the previous "*reproducible research*" emacs configuration, you can easily invoke magit by using `C-x g`.
-[1. Jupyter tips and tricks](#1-jupyter-tips-and-tricks)
-[Creating or importing a notebook](#creating-or-importing-a-notebook)
-[Running R and Python in the same notebook](#running-r-and-python-in-the-same-notebook)
-[Other languages](#other-languages)
-[2. Installing and configuring Jupyter on your computer](#2-installing-and-configuring-jupyter-on-your-computer)
-[2.1 Installing Jupyter](#21-installing-jupyter)
-[2.2 Making sure Jupyter allows you to use R](#22-making-sure-jupyter-allows-you-to-use-r)
-[2.3 Additional tips](#23-additional-tips)
1. Jupyter tips and tricks
==========================
The following [webpage](https://www.dataquest.io/blog/jupyter-notebook-tips-tricks-shortcuts/) lists several Jupyter tricks (in particular, it illustrates many `IPython magic` commands) that should improve your efficiency (note that this blog post is about two years old so some of the tricks may have been integrated in the default behavior of Jupyter now).
Creating or importing a notebook
--------------------------------
Using the Jupyter environment we deployed for this MOOC will allow to easily access any file from your default GitLab project. There are situations however where you may want to play with other notebooks.
Adding a brand new notebook in a given directory
Simply follow the following steps:
1. From the menu: `File -> Open`. You're now in the Jupyter file manager.
2. Navigate to the directory where you want your notebook to be created.
3. Then from the top right button: `New -> Notebook: Python 3`.
4. Give your notebook a name from the menu: `File -> Rename`.
N.B.: If you create a file by doing `File -> New Notebook ->
Python 3`, the new notebook will be created in the current directory. Moving it afterward is possible but a bit cumbersome (you'll have to go through the Jupyter file manager by following the menu `File -> Open`, then select it, `Shut` it `down`, and `Move` and/or `Rename`).
Importing an already existing notebook
If your notebook is already in your GitLab project, then simply synchronize by using the `Git pull` button and use the `File -> Open` menu. Otherwise, imagine, you want to import the [following notebook](https://app-learninglab.inria.fr/gitlab/moocrr-session1/moocrr-reproducibility-study/blob/master/src/Python3/challenger.ipynb) from someone else's repository to re-execute it.
1. Download the file on your computer. E.g., for this [GitLab hosted notebook](https://app-learninglab.inria.fr/gitlab/moocrr-session1/moocrr-reproducibility-study/blob/master/src/Python3/challenger.ipynb), click on `Open raw` (a small `</>` within a document icon) and save (`Ctrl-S` on most browsers) the content (a long Json text file).
2. Open the Jupyter file manager from the menu `File -> Open` and navigate to the directory where you want to upload your notebook.
3. Then from the top right button, `Upload` the previously downloaded notebook and confirm the upload.
4. Open the freshly uploaded notebook through the Jupyter file manager.
Running R and Python in the same notebook
-----------------------------------------
`rpy2` package allows to use both languages in the same notebook by:
1. Loading `rpy2`:
``` python
%load_ext rpy2.ipython
```
2. Using the `%R` Ipython magic:
``` python
%%R
summary(cars)
```
Python objects can then even be passed to R as follows (assuming `df` is a pandas dataframe):
``` python
%%R -i df
plot(df)
```
Note that this `%%R` notation indicates that R should be used for the whole cell but an other possibility is to use `%R` to have a single line of R within a python cell.
Other languages
---------------
Jupyter is not limited to Pytyhon and R. Many other languages are available: [<https://github.com/jupyter/jupyter/wiki/Jupyter-kernels>](https://github.com/jupyter/jupyter/wiki/Jupyter-kernels), including non-free languages like SAS, Mathematica, Matlab... Note that the maturity of these kernels differs widely.
None of these other languages have been deployed in the context of our MOOC but you may want to read the next sections to learn how to set up your own Jupyter on your computer and benefit from these extensions.
Since the question was asked several times, if you really need to stay with SAS, you should know that SAS can be used within Jupyter using either the [Python SASKernel](https://sassoftware.github.io/sas_kernel/) or the [Python SASPy](https://sassoftware.github.io/saspy/) package (step by step explanations about this are given [here](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/documents/tuto_jupyter_windows/tuto_jupyter_windows.md)).
Since proprietary software such as SAS cannot easily be inspected, we discourage its use as it hinders reproducibility by essence. But perfection does not exist anyway and using Jupyter literate programming approach allied with systematic control version and environment control will certainly help anyway.
2. Installing and configuring Jupyter on your computer
In this section, we explain how to set up a Jupyter environment on your own computer similar to the one deployed for this MOOC.
Note that Jupyter notebooks are only a small part of the picture and that Jupyter is now part of a bigger project: [JupyterLab](https://blog.jupyter.org/jupyterlab-is-ready-for-users-5a6f039b8906), which allows you to mix various components (including notebooks) in your browser. In the context of this MOOC, our time frame was too short to benefit from JupyterLab which was still under active development. You may, however, prefer JupyterLab when doing an installation on your own computer.
2.1 Installing Jupyter
----------------------
Follow these instructions if you wish to have a Jupyter environment on your own computer similar to the one we set up for this MOOC.
First, download and install the [latest version of Miniconda](https://conda.io/miniconda.html). We use Miniconda version `4.5.4` and Python version `3.6` on our server.
Miniconda is a light version of Anaconda, which includes Python, the Jupyter Notebook, and other commonly used packages for scientific computing and data science.
Then download the [mooc<sub>rr</sub> environment file](https://gist.github.com/brospars/4671d9013f0d99e1c961482dab533c57) and create the environment using conda:
``` shell
conda env create -f environment.yml
# Windows activate the environment
activate mooc_rr
# Linux and MacOS activate the environment
source activate mooc_rr
# Linux, MacOS and Windows: launch the notebook
jupyter notebook
```
2.2 Making sure Jupyter allows you to use R
-------------------------------------------
The environment described in the last section should include R, but if you proceeded otherwise and only have Python available in Jupyter, you may want to read the following section.
Here is what we had to install on a recent Debian computer to make sure the notebook export via LaTeX works:
``` shell
sudo apt-get install texlive-xetex wkhtmltopdf
```
Obviously, you can convert to html or pdf using the using the `File > Download as > HTML` (or `PDF`) menu option. This can also be done from the command line with the following command:
``` bash
ipython3 nbconvert --to pdf Untitled.ipynb
```
If you want to use a specific style, then the `nbconvert` exporter should be customized. This is discussed and demoed [here](http://markus-beuckelmann.de/blog/customizing-nbconvert-pdf.html). We encourage you to simply read the [doc of nbconvert](https://nbconvert.readthedocs.io/en/latest/).
Instead of going directly through LaTeX and playing too much with the `nbconvert` exporter, an other option consists in exporting to Markdown and playing with [pandoc](https://pandoc.org/). Both approaches work, it's rather a matter of taste.
**Windows**
Download and install MiKTeX from the [MiKTeX webpage](https://miktex.org/download) by choosing the right operating system. You will be prompted to install some specific packages when exporting to pdf.
### • Improving notebook readability
Here are a few extensions that can ease your life:
-[Code folding](https://stackoverflow.com/questions/33159518/collapse-cell-in-jupyter-notebook) to improve readability when browsing the notebook.
``` shell
pip3 install jupyter_contrib_nbextensions
# jupyter contrib nbextension install --user # not done yet
```
-[Hiding code](https://github.com/kirbs-/hide_code) to improve readability when exporting.
``` bash
sudo pip3 install hide_code
sudo jupyter-nbextension install --py hide_code
jupyter-nbextension enable --py hide_code
jupyter-serverextension enable --py hide_code
```
### • Interacting with GitLab and GitHub
To ease your experience, we added pull/push buttons that allow you to commit and sync with GitLab. This development was specific to the MOOC but inspired from a previous [proof of concept](https://github.com/Lab41/sunny-side-up). We have recently discovered that someone else developed about at the same time a [rather generic version of this Jupyter plugin](https://github.com/sat28/githubcommit). Otherwise, remember that it is very easy to insert a shell cell in Jupyter in which you can easily issue git commands. This is how we work most of the time.
This being said, you may have noticed that Jupyter keeps a perfect track of the sequence in which cells have been run by updating the "output index". This is a very good property from the reproducibility point of view but depending on your usage, you may find it a bit painful when committing. Some people have thus developed [specific git hooks](https://gist.github.com/pbugnion/ea2797393033b54674af) to ignore these numbers when committing Jupyter notebooks. There is a long an interesting discussion about various options on [StackOverflow](https://stackoverflow.com/questions/18734739/using-ipython-notebooks-under-version-control).
For those who use [JupyterLab](https://blog.jupyter.org/jupyterlab-is-ready-for-users-5a6f039b8906) rather than the plain Jupyter, a specific [JupyterLab git plugin](https://github.com/jupyterlab/jupyterlab-git) has been developed to offer a nice version control experience.
Since a few years, we systematically require any or our students to have a laboratory notebook in org-mode. Most of the time, they start in private repositories but often end up being fully opened. Here are a few ones:
- Luka Stanisic (a former PhD student advised by Arnaud Legrand) starting using this methodology during his Msc and developed further throughout his PhD. Part of his [PhD thesis](http://mescal.imag.fr/membres/luka.stanisic/thesis/thesis.pdf) was actually about designing a methodology for reproducible experiments in large scale distributed systems. You may want to have a look at [his postdoc LabBook](http://starpu-simgrid.gforge.inria.fr/) and to the [report of Léo Villeveygoux](https://framagit.org/lvgx/pfe/blob/master/doc/labbook.org) whom he advised.
- Tom Cornebize is currently a PhD student advised by Arnaud Legrand and during his MSc, he also heavily [loged his activity on Github](https://github.com/Ezibenroc/simulating_mpi_applications_at_scale).
-[Lucas Schnorr](https://github.com/schnorr)'s students usually also maintain their journal in a very nice way: [Tais Bellini's BSc.](https://github.com/taisbellini/aiyra/blob/master/LabBook.org), [Arthur Krause's LabBook](https://github.com/mittmann/hpc/blob/master/LabBook.org), [Luca Nesi's LabBook](http://www.inf.ufrgs.br/~llnesi/memory_report/MemoryReport.html).
-[Martin Quinson](https://people.irisa.fr/Martin.Quinson/Research/Students/Methodo/)'s students also follow such conventions:
- Ezequiel Torti Lopez, M2R 2014. [Report](https://github.com/mquinson/simgrid-simpar/blob/master/report.org), with both the data provenance and the data analysis included in the appendix.
- Gabriel Corona, engineer on SimGrid, 2015-2016. [Journal](https://github.com/randomstuff/simgrid-journal/blob/master/journal.org), [Blog (findings)](http://www.gabriel.urdhr.fr/tags/simgrid/).
- Matthieu Nicolas, engineer on PLM, 2014-2016, [Journal](https://github.com/MatthieuNICOLAS/PLM-reporting/blob/master/activity-report.org).
Org-mode is obviously not the only option and many of our students use am mixture of org-mode, rstudio and jupyter depending on what is more convenient.
How to report efficiently (by Martin Quinson)
=============================================
My friend Martin has gathered \[\[<https://people.irisa.fr/Martin.Quinson/Research/Students/Methodo/>\]\[an excellent compendium of information and references on his webpage to explain his students what he expects from them\]\]. **I'll therefore simply paraphrase him here** with the most important aspects related to reporting but feel free to read [the original version](https://people.irisa.fr/Martin.Quinson/Research/Students/Methodo/):
Reporting
---------
I ask you to write a little reporting regularly. Depending on the situation, it may be every day, every week or every month. In any case, your reporting is very important for the following reasons:
- It forces you to think about what you are doing, which may help you to unblock your problem by your own. Writing down the problems in a clear way is often sufficient to see the solution appearing.
- It helps me following your progress even between the meetings. I cannot unblock you if I don't detect that you are on a wrong lead or otherwise blocked.
- It keeps a track of the steps in your work. That's good for the day where you want to write your final report (even if a final report should never be presented in the chronological order). That's good for the next after you who will be supposed to continue you effort, or to build upon it.
- That person may be yourself (if you go for a PhD program), another intern, myself or even someone else on the Internet: that's what we call Open Science, an effort where everyone can build upon the scientific work of everyone.
I want you to write your reporting in an org file (yep, you don't have a choice here). \[..\]
Reporting Logistics
-------------------
Once you're setup with all software installed and somehow configured, you need to create a reporting file in a place where I can see it and where it won't get lost if your disk crashes or something. Open a dedicated git repository (on github, gitorious, gitlab, ...) for that. After your internship, your report should be archived directly in the source tree of the software that you are working on, if any. But having your reporting located in the source tree may complicate things during your work.
Yes, it means that your file will be public at some point, but that's why we call it "Open Science", after all. Also, you should write it in English if possible. The part of your reporting that is called "Journal" (see below) may be written in French if you are more efficient this way but the rest must be in English. Don't make your tone too formal because the file is public. Make it efficient. Nobody will ever blame you for the work you did during an internship a long time ago. If you really want, we can even make this file anonymous. Just speak to me.
You want to write your reporting before leaving work. Weekly reporting should be written on Friday, one or two hours before leaving. That's the best solution to have a nice week end without thinking about work, and still lose no information that you would need on Monday morning.
Reporting Document Organization
-------------------------------
Your reporting document should have four main parts:
Findings
This section summarizes the general information that you gathered during your work. It is empty at the beginning of your internship, and gets fleshed with the important things that you find on your way. That's where bibliographical information go, for example. But that's definitely not where TODO notes go (see below).
Development
This section presents the technical sides of your work. Don't write anything in there yet. Put it all in the Journal part for now.
Journal
Describe the day-to-day work done for each period (day, week or month) of your internship. That's the most important part of your reporting, and we come back to it below.
Conclusion
That's what you write in the next week of your internship. You can see it as a letter to the next guy, explaining the current state of your work, a few words about its technical organization, and what should be done next on that topic. Keep this part highly technical, the overall organization of your internship will be seen in your final report.
The Journal part is the only part that you may write in French on need. You want to add one subsection per period to your journal. Don't make it too long, or you would waste time writing long texts that very few will ever read. Don't make it too short or it will be impossible to understand it on Monday morning (or three months after). Finding the good balance is sometimes difficult, but I will provide feedback on your first entries, so don't worry.
Each of section describing a period should contain three subsubsections:
Things done
a few words about what you've done. Something like 2 or 4 items with a few words describing what you've done. You can omit the title of that section and put the items directly in the upper section (see the example below).
Blocking points and questions
try to explain clearly the things that block you or slow you down. If you found the solution already, then it should be part of the previous subsection (but you should say a few words nevertheless). Also ask every question that you may have for me in that section. If the question are personal (e.g., about the logistics of your internship such as salary or so), please prefer emails that are not publicly visible. If this section is empty for a given period, skip it all together (no empty subsubsections).
Planned work
A few items about what you plan to work on during the next period.
A template of reporting file is given at the end of this section. This is just a strong advice: If you really feel better with another file organization, then give it a try for one period, and ask for my feedback. I can adapt, and I do not pretend that my advice is the definitive answer. It's just the result of my experience so far.
Notice how TODO items are written: they are given as items in the Planned work sections of the journal. As explained in the [documentation](http://orgmode.org/manual/Checkboxes.html), you simply have to write "\[\]" in front of items that you plan to do in the future.
You should add a \[1/\] on the "Planned work" line, so that emacs keeps track of what is done and what is still to do. Once they are done, you type C-c C-C on their lines to change the blank box \[\] into a checked box \[X\]. Also, the \[1/\] will be changed to denote the amount of work that is still to be done.
At any point, you can see all ongoing TODO items with the following keystrokes: "C-c / t". More information on TODOs in orgmode's [documentation](http://orgmode.org/manual/TODO-basics.html). The important thing here is that most TODO items must only be written in the *Journal* part (so that we know when they occurred).
**Do not edit past entries of your journal**, unless you have very good reasons. If you must, make sure that you don't lose information about the path that you took (remember the Open Science thingy). You should always **add** information to past entries, such as:
``` shell
- *edit* This hypothesis does not hold; see the entry of [the day where you found it] for more information.
```
The only exception are TODO entries, that should clearly be rewritten to DONE entries. If you need to adapt your TODO entry (because the initial goal was poorly stated or otherwise), change the initial entry from TODO to CANCELED (or check the box after stating in a subitem that it was not done but canceled, and why), and create a new TODO entry in the current period section.
``` example
* Introduction
This file contains the reporting for my beloved internship done on
this topic on that year. For now, just add the official title of
your internship (check the convention signed between your
university and my lab). After a few weeks, once you really
understand your internship, you should write a few paragraphs about
the context, problem and motivation of your work, with some
-[Examples from the Video](#examples-from-the-video)
-[Other examples](#other-examples)
Examples from the Video
=======================
In the MOOC video, I quickly demo how org-mode can be used in various contexts. Here are the (sometimes trimmed) corresponding org-files. These documents depend on many other external data files and are not meant to lead to reproducible documents but it will give you an idea of how it can be organized:
1.[journal.org](journal.org): an excerpt (I've only left a few code samples and links to some resources on R, Stats, ...) from my own journal. This is a personal document where everything (meeting notes, hacking, random thoughts, ...) goes by default. Entries are created with the `C-c c` shortcut.
2.[labbook<sub>single</sub>.org](labbook_single.org): this is an excerpt from the laboratory notebook [Tom Cornebize](https://cornebize.net/) wrote during his Master thesis internship under my supervision. This a personal labbook. I consider this notebook to be excellent and was the ideal level of details for us to communicate without any ambiguity and for him to move forward with confidence.
3.[paper.org](paper.org): this is an ongoing paper based on the previous labbook of Tom Cornebize. As such it is not reproducible as there are hardcoded paths and uncleaned dependencies but writing it from the labbook was super easy as we just had to cut and paste the parts we needed. What may be interesting is the organization and the org tricks to export to the right LaTeX style. As you may notice, in the end of the document, there is a commented section with emacs commands that are automatically executed when opening the file. It is an effective way to depend less on the `.emacs/init.el` which is generally customized by everyone.
4.[labbook<sub>several</sub>.org](labbook_several.org): this is a labbook for a specific project shared by several persons. As a consequence it starts with information about installation, common scripts, has section with notes about all our meetings, a section with information about experiments and an other one about analysis. Entries could have been labeled by who wrote them but there were only a few of us and this information was available in git so we did not bother. In such labbook, it is common to find annotations indicating that such experiment was `:FLAWED:` as it had some issues.
5.[technical<sub>report</sub>.org](technical_report.org): this is a short technical document I wrote after a colleague sent me a PDF describing an experiment he was conducting and asked me about how reproducible I felt it was. It turned out I had to cut and paste the C code from the PDF, then remove all the line numbers and fix syntax, etc. Obviously I got quite different performance results but writing everything in org-mode made it very easy to generate both HTML and PDF and to explicitly explain how the measurements were done.
Other examples
==============
Here are a few links to other kind of examples:
- Slides: all my slides for a series of lectures is available here: <https://github.com/alegrand/SMPE>. Here is a [typical source](https://raw.githubusercontent.com/alegrand/SMPE/master/lectures/lecture_central_limit_theorem.org) and the [resulting PDF](https://raw.githubusercontent.com/alegrand/SMPE/master/lectures/lecture_central_limit_theorem.pdf)
- Lucas Schnorr, a colleague, maintains:
- a set of templates for various computer science journals/conferences: [IEEE](https://github.com/schnorr/ieeeorg), [Wiley](https://github.com/schnorr/wileyorg), [ACM](https://github.com/schnorr/acmorg), [LNCS](https://github.com/schnorr/llncsorg)
- his lecture on programming languages for undergrads: <https://github.com/schnorr/mlp/tree/master/conteudo>
- John Kitchin is an expert org-mode user and he maintains a very interesting [blog with many interesting tips](http://kitchingroup.cheme.cmu.edu/blog/). You may want to check this [seminar he gave at SciPy](https://www.youtube.com/watch?v=IsSMs-4GlT8&list=FLQp2VLAOlvq142YN3JO3y8w&app=desktop).