Voici un aperçu rapide de la syntaxe Markdown repris d'une [présentation de Github](https://guides.github.com/features/mastering-markdown/) ainsi que de celles d'[Archer Reilly](http://csrgxtu.github.io/2015/03/20/Writing-Mathematic-Fomulars-in-Markdown/).
Table des matières<span class="tag" data-tag-name="TOC"></span>
To print some text with python, you should use the `print()` function.
```
print("Hello world!")
```
````
Écrire des Maths
================
Il est possible d'écrire des formules en Markdown, soit en mode **inline** soit en mode **displayed formulas**. Dans le premier cas, les formules sont inclues directement à l'intérieur du paragraphe courant alors que dans le second, elles apparaissent centrées et mises en exergue.
Le formatage de la formule est légèrement différent dans les deux cas car pour qu'une formule s'affiche joliment sur une seule ligne, il faut la "tasser" un peu plus que lorsqu'elle est mise en valeur.
Pour écrire une formule en mode **inline**, il faut la délimiter par un `$` (du coup, pour écrire le symbole dollar, il faut le préfixer par un backslash, comme ceci: `\$`) alors que pour écrire en mode **displayed**, il faut la délimiter par un `$$`. Un petit exemple valant mieux qu'un long discours, voici concrètement comment cela fonctionne:
``` example
Cette expression $\sum_{i=1}^n X_i$ est inlinée.
```
Cette expression $\\sum\_{i=1}^n X\_i$ est inlinée.
``` example
Cette expression est mise en valeur:
$$\sum_{i=1}^n X_i$$
```
Cette expression est mise en valeur:
$$\\sum\_{i=1}^n X\_i$$
Nous vous présentons par la suite une sélection de symboles et de commandes courantes. En fait, à peu près tout ce qui est classique dans le langage LaTeX peut être utilisé pourvu que vous délimitiez bien avec un `$`. Pour d'autres exemples plus complets jetez un coup d'œil à ces [exemples de James H. Steiger](http://www.statpower.net/Content/310/R%2520Stuff/SampleMarkdown.html).
Tout d'abord, pour aller plus loin avec `markdown` et ses extensions / ramifications :
- Le didacticiel « [Élaboration et conversion de documents avec Markdown et Pandoc](https://enacit1.epfl.ch/markdown-pandoc/) » de Jean-Daniel Bonjour (EPFL), précis, complet, concis, en français ; un vrai bonheur !
- L'article [Markdown](https://en.wikipedia.org/wiki/Markdown#Example) de wikipedia en anglais contient un bon pense-bête sur la syntaxe `markdown`.
- Github propose un court et efficace didacticiel (en anglais) : [Mastering Markdown](https://guides.github.com/features/mastering-markdown/).
Comme nous l'illustrons dans la « film d'écran » (*screencast*), l'éditeur de texte des dépôts `github` et `gitlab` permet d'interpréter / transformer à la demande un fichier `mardown` en un fichier `html`. C'est à la fois agréable et pratique, mais ce n'est pas une solution pour une utilisation quotidienne de `markdown`, pour cela, il est plus efficace d'éditer son texte, avec un éditeur de texte, sur son ordinateur, avant de « l'exporter » dans un format comme `html`, `pdf`, `docx`, `epub`, etc. Il existe des éditeurs plus ou moins spécialisés pour `markdown`, certains sont indiqués sur la page [Editors](https://github.com/jgm/pandoc/wiki/Pandoc-Extras#editors) du site de `pandoc`, mais nous préconisons clairement l'emploi d'un éditeur de texte « généraliste » capable de reconnaître la syntaxe `markdown`. Nous en avons indiqué en début de séquence et on pourra trouver des informations complémentaires dans la section [Quelques éditeurs adaptés à l'édition Markdown](https://enacit1.epfl.ch/markdown-pandoc/#editeurs_markdown) du didacticiel de Jean-Daniel Bonjour.
Pour convertir un fichier `markdown` en un format « arbitraire », la solution à ce jour la plus complète est [Pandoc](http://pandoc.org/), logiciel développé par John MacFarlane, un philosophe de Berkeley (le site [github](https://github.com/jgm/pandoc)). En plus du site de `Pandoc`, le didacticiel de J.-D. Bonjour donne de nombreuses explications sur comment installer et utiliser `pandoc` dans la section [Utilisation du convertisseur Pandoc](https://enacit1.epfl.ch/markdown-pandoc/#commande_pandoc). Comme `pandoc` -- écrit en Haskell -- peut être parfois un peu difficile à installer, nous indiquons maintenant quelques solutions alternatives :
- Des sites comme <http://www.markdowntopdf.com/> et <http://markdown2pdf.com/> permettent de convertir en ligne un fichier `markdown` en un fichier `pdf`.
- Le projet [CommonMark](http://commonmark.org/) propose, en plus d'une spécifications plus rigoureuse de la syntaxe `markdown`, des convertisseurs `markdown` → `html` / `LaTeX` (et plus) écris en `C` et en `JavaScript` (<https://github.com/CommonMark/CommonMark>).
- Le site de [John Gruber](https://daringfireball.net/projects/markdown/), le créateur de `markdown`, fournit un convertisseur `markdown` → `html` écrit en `perl`.
-[MultiMarkdown](http://fletcherpenney.net/multimarkdown/) est une autre extension de `markdown` qui vient avec son convertisseur `markdown` → `html` écrit en `C`.
-[grip](https://github.com/joeyespo/grip) est un serveur écrit en `python` qui permet de convertir et visualiser à la volée des fichiers `markdown` avec son navigateur (très utile pour éviter d'avoir à faire des « commits » en grande quantité lorsqu'on écrit de tels fichiers pour un dépôt `github` ou `gitlab`).
La conversion en `pdf` passe toujours par [LaTeX](https://fr.wikipedia.org/wiki/LaTeX) ce qui nécessite d'avoir une version complète et à jour de ce logiciel sur sa machine.
Dans la petite démonstration, nous montrons comment générer un fichier `docx` à partir d'un fichier `md` avec `Pandoc` et nous soulignons qu'il est alors possible d'utiliser un traitement de texte comme `LibreOffice` pour modifier le fichier obtenu. Il est clair que si des modifications sont apportées au `docx` elle en seront pas (automatiquement) propagées au `md`. Il faudra utiliser `Pandoc` pour cela et effectuer une conversion de `docx` vers `md` (et seules les éléments du format `docx` qui existent en `md` seront conservés).
Une stratégie qui est souvent employée et qui fonctionne bien en pratique consiste à faire le gros du travail de rédaction d'un article ou d'un mémoire en `Markdown`. La rédaction terminée, le fichier est exporté au format `docx` (ou `LaTeX`) et des ajustements de mise en page sont alors effectués avec un logiciel de traitement de texte (ou un éditeur `LaTeX`).
**Disclaimer:** The two sections *A simple "reproducible research" emacs
**Disclaimer:** The two sections *A simple "reproducible research" emacs configuration* and *A stub of replicable article* explain how to set up emacs/org-mode for this MOOC. These are very important sections in the context of this MOOC. **These sections are illustrated in two out of the [three video tutorials of this sequence](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4), and****which you really should follow carefully**. **Otherwise, you may have trouble doing the exercises later on**. Likewise, I strongly encourage you to watch the ["emacs and git" video tutorial available at the same place](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4).
configuration* and *A stub of replicable article* explain how to set up
emacs/org-mode for this MOOC. These are very important sections in the
The next section provides information on how to install emacs.
context of this MOOC. **These sections are illustrated in two out of the
[three video tutorials of this
Table of Contents<span class="tag" data-tag-name="TOC"></span>
We provide here only instructions for Debian-based distributions. Feel
We provide here only instructions for Debian-based distributions. Feel free to contribute to this document to provide up-to-date information for other distributions (e.g.n redhat, fedora).
free to contribute to this document to provide up-to-date information
for other distributions (e.g.n redhat, fedora).
Today, the stable versions of the most common distributions provide
Today, the stable versions of the most common distributions provide recent enough versions of emacs and org-mode:
recent enough versions of emacs and org-mode:
- Debian (stretch) ships with
- Debian (stretch) ships with [emacs 25.1](https://packages.debian.org/stretch/emacs25) and [org-mode 9.0.3](https://packages.debian.org/stretch/org-mode)
[emacs 25.1](https://packages.debian.org/stretch/emacs25) and
- Ubuntu (bionic 18.04) ships with [emacs 25.2](https://packages.ubuntu.com/bionic/emacs25) and [org-mode 9.1.6](https://packages.ubuntu.com/bionic/org-mode)
Org mode version 9.1.11 (9.1.11-dist @ /usr/share/emacs/25.2/site-lisp/elpa/org-9.1.11/)
Org mode version 9.1.11 (9.1.11-dist @ /usr/share/emacs/25.2/site-lisp/elpa/org-9.1.11/)
```
```
The version numbers you get will depend on the distribution you are
The version numbers you get will depend on the distribution you are running. **You really want to make sure you do not rely on org-mode 8**, which is now deprecated.
running. <spanclass="underline">You really want to make sure you do not
rely on org-mode 8</span>, which is now deprecated.
## macOS
macOS
-----
**Note:** macOS comes with a prehistoric command-line-only version of
**Note:** macOS comes with a prehistoric command-line-only version of Emacs located at `/usr/bin/emacs`. It's best to forget about it.
Emacs located at `/usr/bin/emacs`. It's best to forget about it.
-**Option 1**: Install the `.dmg` file from [Vincent
Goulet](http://vgoulet.act.ulaval.ca/):
<https://vigou3.gitlab.io/emacs-modified-macos/>. It ships with
recent versions:
-**Option 1**: Install the `.dmg` file from [Vincent Goulet](http://vgoulet.act.ulaval.ca/): [<https://vigou3.gitlab.io/emacs-modified-macos/>](https://vigou3.gitlab.io/emacs-modified-macos/). It ships with recent versions:
- Emacs 26.1
- Emacs 26.1
- Org-mode 9.1.13
- Org-mode 9.1.13
- ESS 17.11
- ESS 17.11
If you install this version of Emacs, or in fact any other version
If you install this version of Emacs, or in fact any other version of Emacs distributed as a clickable application in a `.dmg` file, you must type the full path to the executable if you want to run Emacs from a terminal. For example, if your clickable application is at `/Applications/Emacs.app`, then the executable is at `/Applications/Emacs.app/Contents/MacOS/Emacs`
of Emacs distributed as a clickable application in a `.dmg` file,
you must type the full path to the executable if you want to run
Emacs from a terminal. For example, if your clickable application is
at `/Applications/Emacs.app`, then the executable is at
`/Applications/Emacs.app/Contents/MacOS/Emacs`
-**Option 2**: If you use [Homebrew](https://docs.brew.sh/), do the
-**Option 2**: If you use [Homebrew](https://docs.brew.sh/), do the following:
following:
``` bash
``` bash
brew update
brew update
...
@@ -132,12 +93,7 @@ Emacs located at `/usr/bin/emacs`. It's best to forget about it.
...
@@ -132,12 +93,7 @@ Emacs located at `/usr/bin/emacs`. It's best to forget about it.
brew install ess
brew install ess
```
```
This provides an `emacs`command for use from the command line, plus
This provides an `emacs` command for use from the command line, plus a clickable application at `Cellar/emacs/26.1_1/Emacs.app` inside your Homebrew directory. If you installed Homebrew at the default location `/usr/local`, then this is `/usr/local/Cellar/emacs/26.1_1/Emacs.app`. If you installed Homebrew on an account with administrator privileges, you can add
a clickable application at `Cellar/emacs/26.1_1/Emacs.app` inside
your Homebrew directory. If you installed Homebrew at the default
location `/usr/local`, then this is
`/usr/local/Cellar/emacs/26.1_1/Emacs.app`. If you installed
Homebrew on an account with administrator privileges, you can add
``` bash
``` bash
brew linkapps emacs
brew linkapps emacs
...
@@ -145,38 +101,24 @@ Emacs located at `/usr/bin/emacs`. It's best to forget about it.
...
@@ -145,38 +101,24 @@ Emacs located at `/usr/bin/emacs`. It's best to forget about it.
in order to make Emacs accessible directly from `/Applications`.
in order to make Emacs accessible directly from `/Applications`.
## Windows
Windows
-------
Install the `.exe` file from [Vincent
Install the `.exe` file from [Vincent Goulet](http://vgoulet.act.ulaval.ca/): [<https://vigou3.gitlab.io/emacs-modified-windows/>](https://vigou3.gitlab.io/emacs-modified-windows/). It ships with recent versions:
Goulet](http://vgoulet.act.ulaval.ca/):
<https://vigou3.gitlab.io/emacs-modified-windows/>. It ships with recent
versions:
- Emacs 26.1
- Emacs 26.1
- Org-mode 9.1.13
- Org-mode 9.1.13
- ESS 17.11
- ESS 17.11
### Directory naming conventions
### Directory naming conventions
In the following instructions, we refer to your home directory through
In the following instructions, we refer to your home directory through the (UNIX) `~/` notation. On Windows, your home directory should be something like `C:\Users\yourname`. Therefore, whenever we mention the `~/org/` (resp. the `~/.emacs.d/`) directory this means we are referring to `C:\Users\yourname\org` (resp. `C:\Users\yourname\.emacs.d\`).
the (UNIX)`~/` notation. On Windows, your home directory should be
something like `C:\Users\yourname`. Therefore, whenever we mention the
`~/org/`(resp. the `~/.emacs.d/`) directory this means we are referring
to `C:\Users\yourname\org`(resp. `C:\Users\yourname\.emacs.d\`).
### Making R and Python available to the console
### Making R and Python available to the console
When running a command, Windows will look for the command in the
When running a command, Windows will look for the command in the directories indicated in the `PATH` environment variable. If none of these directories contains the command, Windows will stop and indicate the command does not exist. To make sure R (which may be in something like `C:/Program Files/R/R-3.5.1/bin/x64/`) and Python (which may be in something like `C:/Program Files/Python/Python37/`) can easily be run from Emacs, you should thus configure the `PATH` variable accordingly.
directories indicated in the `PATH` environment variable. If none of
these directories contains the command, Windows will stop and indicate
the command does not exist. To make sure R (which may be in something
like `C:/Program Files/R/R-3.5.1/bin/x64/`) and Python (which may be in
something like `C:/Program Files/Python/Python37/`) can easily be run
from Emacs, you should thus configure the `PATH` variable accordingly.
This requires to go through the "Environment Variable" editor as
This requires to go through the "Environment Variable" editor as explained [here](http://sametmax.com/ajouter-un-chemin-a-la-variable-denvironnement-path-sous-windows/).
Then you will want to deactivate interactive plots in matplotlib. To
Then you will want to deactivate interactive plots in matplotlib. To this end, you first need to know where the matplotlib configuration is located. Open a python console the type the following code:
this end, you first need to know where the matplotlib configuration is
located. Open a python console the type the following code:
Open the `matplotlibrc` file and modify the line starting with `backend`
Open the `matplotlibrc` file and modify the line starting with `backend` to make it `backend : Agg`.
to make it `backend : Agg`.
## All platforms: pretty code in HTML export
All platforms: pretty code in HTML export
-----------------------------------------
To have code pretty printing when exporting to HTML, you should install
To have code pretty printing when exporting to HTML, you should install the `htmlize` package, which is done by opening emacs and typing the following command:
the `htmlize` package, which is done by opening emacs and typing the
following
command:
``` example
``` example
M-x package-install RET htmlize RET # where M-x means pressing the "Esc" key then the "x" key
M-x package-install RET htmlize RET # where M-x means pressing the "Esc" key then the "x" key
```
```
# A simple "*reproducible research*" emacs configuration
A simple "*reproducible research*" emacs configuration
This section is illustrated in a [video tutorial](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4) (/"Mise en place Emacs/Orgmode"/ in French). Watching it before following the instructions given in this section may help.
(*"Mise en place Emacs/Orgmode"* in French). Watching it before
following the instructions given in this section may help.
Emacs comes with very basic default configuration and it appears like
Emacs comes with very basic default configuration and it appears like everyone has its own taste. You will for example find [here](https://www.emacswiki.org/emacs/StarterKits) several default Emacs configurations that reflect the preferences of their creators. Likewise the configuration of Org-Mode is incredibly flexible (see for example [the org-mode website](https://orgmode.org/worg/org-configs/index.html) for more references). In the context of this MOOC, we propose you a relatively minimalist one that is rather "*reproducible research*" oriented by adding a few org-mode specific configurations.
everyone has its own taste. You will for example find
[here](https://www.emacswiki.org/emacs/StarterKits) several default
Emacs configurations that reflect the preferences of their creators.
Likewise the configuration of Org-Mode is incredibly flexible (see for
example [the org-mode
website](https://orgmode.org/worg/org-configs/index.html) for more
references). In the context of this MOOC, we propose you a relatively
minimalist one that is rather "*reproducible research*" oriented by
adding a few org-mode specific configurations.
## Step 0: Backup and download our configuration
Step 0: Backup and download our configuration
---------------------------------------------
The procedure we propose will wipe your already existing custom Emacs
The procedure we propose will wipe your already existing custom Emacs configuration if you have one. **You should thus beforehand make a backup** of `~/.emacs` and of `~/.emacs.d/init.el` (if these files exist).
configuration if you have one. **You should thus beforehand make a
backup** of `~/.emacs` and of `~/.emacs.d/init.el` (if these files
exist).
Then download [this
Then download [this archive](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/rr_org_archive.tgz) and uncompress it. It contains the following files and we will refer to them in the following:
and uncompress it. It contains the following files and we will refer to
them in the following:
``` example
``` example
rr_org/init.el
rr_org/init.el
rr_org/journal.org
rr_org/journal.org
```
```
Alternatively, [the files you are looking for are available
Alternatively, [the files you are looking for are available here](rr_org/).
here](rr_org/).
## Step 1: Prepare your journal
Step 1: Prepare your journal
----------------------------
Create an `org/` directory in the top of your home:
Create an `org/` directory in the top of your home:
...
@@ -259,42 +180,33 @@ Create an `org/` directory in the top of your home:
...
@@ -259,42 +180,33 @@ Create an `org/` directory in the top of your home:
mkdir -p ~/org/
mkdir -p ~/org/
```
```
Then copy `rr_org/journal.org` file in your `~/org/` directory. This
Then copy `rr_org/journal.org` file in your `~/org/` directory. This file will be your laboratory notebook and all the notes you will capture with `C-c c` will go automatically go in this file. The first entry of this notebook is populated with [many Emacs shortcuts](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/rr_org/journal.org) that you should give a try.
file will be your laboratory notebook and all the notes you will capture
with `C-c c` will go automatically go in this file. The first entry of
Copy `rr_org/init.el` in your `~/.emacs.d/` directory.
Copy `rr_org/init.el` in your `~/.emacs.d/` directory.
Alternatively, if you do not want to mess with your already existing
Alternatively, if you do not want to mess with your already existing emacs configuration, you may launch emacs with this specific configuration with the following command: `emacs -q -l rr_org/init.el`.
emacs configuration, you may launch emacs with this specific
configuration with the following command: `emacs -q -l rr_org/init.el`.
## Step 3: Adapt the configuration to your specific needs if required
Step 3: Adapt the configuration to your specific needs if required
Open a new instance of Emacs and open a `foo.org` file. Copy the following lines in this file:
``` example
``` example
#+begin_src shell :session foo :results output :exports both
#+begin_src shell :session foo :results output :exports both
...
@@ -302,50 +214,25 @@ ls -la # or dir under windows
...
@@ -302,50 +214,25 @@ ls -la # or dir under windows
#+end_src
#+end_src
```
```
Put your cursor inside this code block and execute it with the following
Put your cursor inside this code block and execute it with the following command: `C-c C-c` (If you are not familiar with Emacs commands, this one means '`Ctrl + C`' twice)
command: `C-c C-c` (If you are not familiar with Emacs commands, this
one means '`Ctrl + C`' twice)
A `#+RESULTS:` block with the result of the command should appear if it worked.
A `#+RESULTS:` block with the result of the command should appear if it
In the video, we already have demonstrated the main features and shortcuts of emacs/org-mode that will help you maintain a document and benefit from literate programming. The list of features and shortcuts is demonstrated in the [first entry of your labbook](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/rr_org/journal.org).
worked.
Step 5: Open and play with your journal:
In the video, we already have demonstrated the main features and
----------------------------------------
shortcuts of emacs/org-mode that will help you maintain a document and
benefit from literate programming. The list of features and shortcuts is
In step 1, you were told to create an journal in `~org/journal.org`. First you probably want to make sure this file is stored in a version control system like git. We leave it up to you to set this up but if you have any trouble, feel free to ask on the FUN forums.
This section is illustrated in a [video tutorial](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4) (/"Écrire un article réplicable avec Emacs/Orgmode"/ in French). Watching it before following the instructions given in this section may help.
In step 1, you were told to create an journal in `~org/journal.org`.
First you probably want to make sure this file is stored in a version
Remember, you need a working LaTeX and R environment. If you can't open a terminal and run the commands `R`, `pdflatex`, and `python`, you will not be able to generate this document. When being compiled, the article downloads the corresponding LaTeX packages so you also need to have a working `wget` command (alternatively, it uses `curl`). Once downloaded, you may still read the source (<https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/replicable_article/article.org>) and understand how it works though.
control system like git. We leave it up to you to set this up but if you
have any trouble, feel free to ask on the FUN forums.
Download the following [archive](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/replicable_article.tgz), uncompress it and simply `make` to generate the article. You should then be able to open the [resulting article](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/replicable_article/article.pdf). This is summarized in the following command:
@@ -354,99 +241,64 @@ tar zxf replicable_article.tgz; cd replicable_article; make ; evince article.pdf
...
@@ -354,99 +241,64 @@ tar zxf replicable_article.tgz; cd replicable_article; make ; evince article.pdf
**Possible issues**:
**Possible issues**:
- If the `make` command fails (especially on Mac), it may be because
- If the `make` command fails (especially on Mac), it may be because Emacs or something else is not correctly installed. In that case, open the article directly with the following command:
Emacs or something else is not correctly installed. In that case,
and export it to pdf with the following shortcut: `C-c C-e l o`
and export it to pdf with the following shortcut: `C-c C-e l o`
- If it still doesn't work and emacs complains about not finding ESS, it may be because you installed ESS in your home instead of system-wide. In that case, try to remove the `-q` in the previous command line to load your personal emacs configuration.
- If it still doesn't work and emacs complains about not finding ESS,
Finally, when you'll be tired of always re-executing all the source code when exporting, just look for the following line in [article.org](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/replicable_article/article.org):
it may be because you installed ESS in your home instead of
system-wide. In that case, try to remove the `-q` in the previous
command line to load your personal emacs configuration.
Finally, when you'll be tired of always re-executing all the source code
when exporting, just look for the following line in
If you remove the `# ` in the beginning of the line, it will not be a
If you remove the `# ` in the beginning of the line, it will not be a comment anymore and will indicate org-mode to stop evaluating every chunk of code when exporting.
comment anymore and will indicate org-mode to stop evaluating every
chunk of code when exporting.
# Emacs tips and tricks
Emacs tips and tricks
=====================
## Cheat-sheets
Cheat-sheets
------------
Learning Emacs and Org-Mode can be difficult as there is an inordinate
Learning Emacs and Org-Mode can be difficult as there is an inordinate amount of shortcuts. Many people have thus come up with cheat-sheats. Here is a selection in case it helps:
amount of shortcuts. Many people have thus come up with cheat-sheats.
Here is a selection in case it helps:
### Emacs
### Emacs
-[Common and step-by-step Emacs shortcuts for our *reproducible
- [Common and step-by-step Emacs shortcuts for our *reproducible research* configuration](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/rr_org/journal.org)
research*
- [The official GNU emacs refcard](https://www.gnu.org/software/emacs/refcards/pdf/refcard.pdf)
- Two graphical cheat-sheats by Sacha Chua on 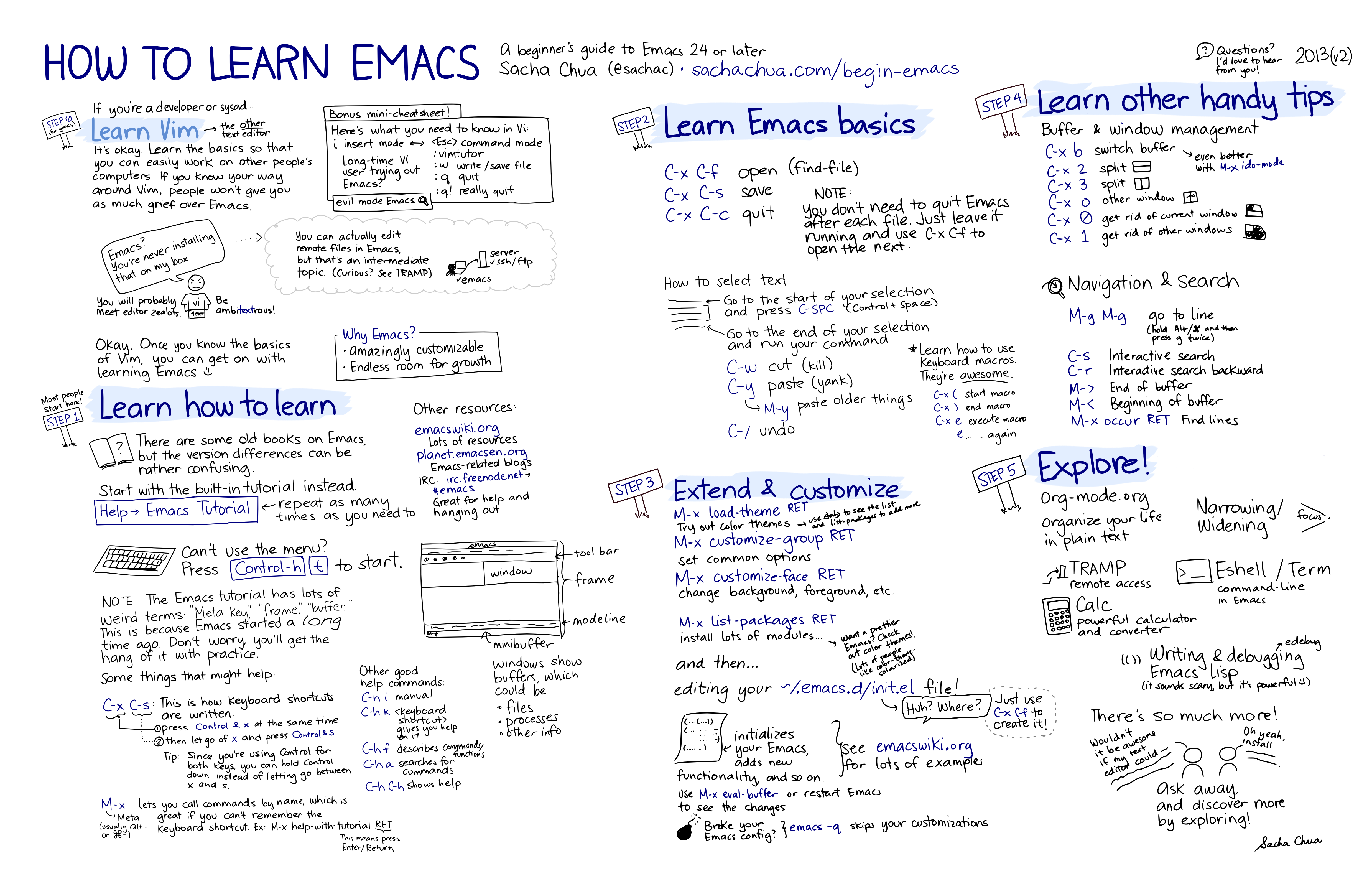 and on .
- [The official description of the org-mode syntax](https://orgmode.org/worg/dev/org-syntax.html) and a [relatively concise description of the org-mode syntax](https://gist.github.com/hoeltgman/3825415).
-[The official org-mode
refcard](https://orgmode.org/worg/orgcard.html)
-[The official description of the org-mode
syntax](https://orgmode.org/worg/dev/org-syntax.html) and a
For those of you who prefer video explanations, here is a [Youtube
For those of you who prefer video explanations, here is a [Youtube channel with many step by step emacs tutorials](https://www.youtube.com/playlist?list=PL9KxKa8NpFxIcNQa9js7dQQIHc81b0-Xg).
[Company-mode](http://company-mode.github.io/) is a text completion
[Company-mode](http://company-mode.github.io/) is a text completion framework for Emacs. It allows to have smart completion in emacs for the most common languages. If you feel this is needed, you should follow the instructions from the official Web page: [<http://company-mode.github.io/>](http://company-mode.github.io/)
framework for Emacs. It allows to have smart completion in emacs for the
most common languages. If you feel this is needed, you should follow the
instructions from the official Web page:
<http://company-mode.github.io/>
### Magit
### Magit
[Magit](https://magit.vc/) is an Emacs interface for Git. Its usage is
[Magit](https://magit.vc/) is an Emacs interface for Git. Its usage is briefly illustrated in the context of this MOOC in a [video tutorial](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/9cfc7500f0ef46d288d2317ec7b037b4) ("*Utilisation Emacs/git*" in French).
briefly illustrated in the context of this MOOC in a [video
It is very powerful and we use it on a daily basis but you should definitely understand what git does behind the scenes beforehand. If you feel this would be useful for you, you should follow [this visual walk-through](https://magit.vc/screenshots/) or [this really short "crash course"](https://www.emacswiki.org/emacs/Magit). If you installed the previous "*reproducible research*" emacs configuration, you can easily invoke magit by using `C-x g`.
("*Utilisation Emacs/git*" in French).
It is very powerful and we use it on a daily basis but you should
Other resources
definitely understand what git does behind the scenes beforehand. If you
---------------
feel this would be useful for you, you should follow [this visual
walk-through](https://magit.vc/screenshots/) or [this really short
"crash course"](https://www.emacswiki.org/emacs/Magit). If you installed
the previous "*reproducible research*" emacs configuration, you can
**This document is particularly important if you follow the RStudio or the Org-Mode path.****If you follow the Jupyter path, it can be ignored at first****as we have closely integrated Jupyter and GitLab in the context of this MOOC.**
So far, you only used git via the web interface from the GitLab we deployed for the MOOC: <https://app-learninglab.inria.fr/gitlab/>
If you access this link from the FUN platform, you do not have to authenticate and you can readily read and modify all your files. This is very convenient but in most cases, you will want to have your own local copy of the repository and you will have to synchronize your local copy with the remote GitLab one. To propagate your modifications, you will obviously have to authenticate yourself on GitLab.
This document describes the software you need to have installed on your machine and how to handle authentication. The "Configuring Git" section is illustrated in a [video tutorial](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/7508aece244548349424dfd61ee3ba85)(in French).
Please read all these instructions carefully, in particular the one on "Configuring your password on GitLab".
Table of Contents<span class="tag" data-tag-name="TOC"></span>
-[Telling Git who you are: Name and Email](#telling-git-who-you-are-name-and-email)
-[Dealing with proxies](#dealing-with-proxies)
-[Getting your default password on GitLab (and possibly changing it)](#getting-your-default-password-on-gitlab-and-possibly-changing-it)
-[Remembering your password locally](#remembering-your-password-locally)
-[Optional: authenticating through SSH](#optional-authenticating-through-ssh)
-[Using Git through the command line to synchronize your local files with Gitlab](#using-git-through-the-command-line-to-synchronize-your-local-files-with-gitlab)
Installing Git
==============
Linux (Debian, Ubuntu)
----------------------
We provide here only instructions for Debian-based distributions. Feel free to contribute to this document to provide up-to-date information for other distributions (e.g., RedHat, Fedora).
Run (as root):
``` bash
apt-get update ; apt-get install git
```
Mac OSX and Windows
-------------------
- Download and install Git from the [Git website](https://git-scm.com/downloads).
- Optional Git clients (should not be needed if you work within RStudio):
-[SourceTree](https://www.sourcetreeapp.com/)
-[GitHub Desktop](https://desktop.github.com/)
> [Apparently](https://github.com/desktop/desktop/issues/852), this works with GitLab and https.
3. Confirm that you have set the Git username correctly:
``` shell
git config --global user.name
git config --global user.email
```
``` example
Mona Lisa
email@example.com
```
Dealing with proxies
--------------------
You may be behind a proxy, in which case you may have trouble cloning or fetching from a remote repository or you may get an error like `unable to access ... Couldn't resolve host ...`
The `proxyPassword` will be stored in plain text (unencrypted) in your `.gitconfig` file, which you may not want. In that case, remove it from the URL and you will be prompted for it every time it is needed.
Getting your default password on GitLab (and possibly changing it)
**Warning (Jupyter users) :** changing your default Gitlab password will prevent you from committing in Jupyter. You will have to do the extra step of changing your `~/.git-credentials` in the Jupyter environment (possibly several times).
1. Get your default password using the [Gitlab credentials retrieval tool](https://app-learninglab.inria.fr/jupyterhub/services/password) as described on the [corresponding resource](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/7508aece244548349424dfd61ee3ba85).

The first long and ugly character sequence is your GitLab login/id. It is easy to find once you are logged on gitlab. The second one however is your password and this webpage is the only place where you can find it. We used the FUN authentification mechanism to propagate your credentials so only you can have access to it. You'll need to use this password when trying to propagate some modifications from your computer to GitLab.
*Note: You have to access this webpage from the FUN platform otherwise you may get a 405 error* *when trying to direcly open <https://app-learninglab.inria.fr/jupyterhub/services/password>.*

2. Access [GitLab](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/5571950188c946e790f06d4bc90fb5f6) from the FUN plateform (click on "Accédez à Gitlab / Access to Gitlab" Button).
*Note: Again, you have to access Gitlab from the FUN platform otherwise you may get a 405 error* /when trying to direcly open <https://app-learninglab.inria.fr/gitlab/users/sign_in/>.
3. Click on the first `Sign in` button (alternatively, you can the login/password you just retrieved and use the second `Sign in` button).

4. If you wish to modify your password, you should go to `Account > Settings > Password` and define your password using the default password you just retrieved. Again, if you use the Jupyter notebooks we have deployed for the MOOC, remember that changing your default Gitlab password will prevent you from committing in Jupyter. You will have to do the extra step of changing your Jupyter `~/.git-credentials` through a Jupyter console (see next section).

Remembering your password locally
---------------------------------
If you clone your repository by simply pasting the GitLab URL, you will be prompted for your login and your password every time you want to propagate your local modifications, which is tedious. This is why you should ask git to remember your login and password as follows
``` shell
git config --global credential.helper cache # remember my password
git config --global credential.helper "cache --timeout=3600"# for one hour at most
```
With this setup, you will be prompted for your password but it will be cached in memory and they will not be asked again before an hour. You may want to read [these instructions](https://stackoverflow.com/questions/5343068/is-there-a-way-to-skip-password-typing-when-using-https-on-github) to better understand how all this works.
If you want your password to be permanently remembered, you should use this command
``` shell
git config credential.helper store
```
Your password will be then stored in a `.git-credentials` file in plain text. On a perfectly secured machine, it may be fine... or not... ;) Use it at your own risk.
Optional: authenticating through SSH
------------------------------------
There are two ways of authenticating and synchronizing your local repository with GitLab: through HTTPS or through SSH. The first one is what was just described and does not require any particular software installation on your machine so this is what I recommend for this MOOC. Yet, I like the second one better (although is may seem a bit more technical), which is why I describe it here. It consists in installing SSH, creating a pair or private/public keys, and uploading your SSH public key on GitLab. This section provides with information on how to do this.
### Installing SSH
1. Linux (Debian, Ubuntu)
We provide here only instructions for debian-based distributions. Feel free to contribute to this document to provide up-to-date information for other distributions (e.g., RedHat, Fedora).
Run (as root):
``` bash
apt-get update ; apt-get install openssh-client
```
2. macOS
You do not have anything to do as it is installed by default.
3. Windows
You should install the [Putty](https://www.ssh.com/ssh/putty/windows/) client. Once it is installed, look for the section on [generating an SSH key](https://www.ssh.com/ssh/putty/windows/puttygen).
### Setting up SSH on GitLab
Here are [all the official explanations on how to set up your SSH key on GitLab](https://docs.gitlab.com/ee/ssh/). Alternatively, you may also want to have a look at this video:
This section describes a generic (through the command line) way to synchronize your local files with Gitlab. You will not need this if you follow the Jupyter path. If you follow the RStudio path, all these operations can be done through RStudio and you may want to read [the corresponding instructions](file:///jump_to_id/1a4f58a1efed437c93a9f5c5f15df428). If you follow the Org-Mode path, all these operations can be done through Magit and you may want to read [the corresponding instructions](file:///jump_to_id/508299f7373449a3939faa5b11462bc4).
Here are other ways to learn Git through the command line:
- The [Software Carpentry git tutorial](http://swcarpentry.github.io/git-novice/)
- The (freely available) Pro Git book ([in English](https://git-scm.com/book/en/v2) or [in French](https://git-scm.com/book/fr/v2)). Reading the first two chapters is enough to get a good start.
-[Learn Git Branching](https://learngitbranching.js.org/) will allow to interactively learn Git and to understand with branches.
Now, let's start!
1. Obtain the repository URL

2. Cloning the repository
``` shell
cd /the/directory/where/you/want/to/clone/your/repository
Now a directory `mooc-rr` has been created on your computer.
3. Inspect the repository
``` shell
cd mooc-rr
ls # (Unix)
dir # (Windows)
```
4. Synchronizing to GitLab
You should indicate which files to track (`git add`) and commit them locally (`git commit`) before they can be transfered (`git push`) to GitLab. The `git status` will indicate you whether files are tracked/modified/committed/...
Let's assume you just created a `fichier.txt` file on the top of the `mooc-rr` directory.
``` shell
git status
```

``` shell
git add fichier.txt
git status
```

``` shell
git commit -m "message commit"
```

``` shell
git status
```

The file can then be transfered to GitLab:
``` shell
git push
```
At this point, git will ask you about your login/password unless you followed the previous *Remembering your password locally* instructions.
N.B.: you will not be allowed to propagate your modifications to GitLab if other modifications (e.g., from someone else) have been propagated in between

5. Synchronizing from Gitlab: to avoid the previous problem, you need to fetch the remote GitLab modifications first and apply them locally.
**Ce document est particulièrement important si vous suivez le parcours RStudio ou Org-Mode. Vous pouvez l’ignorer dans un premier temps si****vous suivez le parcours Jupyter car nous avons étroitement intégré Jupyter et GitLab dans le contexte de ce MOOC.**
Jusqu'à présent, vous avez utilisé Git uniquement via l'interface web du GitLab que nous avons déployée pour le MOOC : [<https://app-learninglab.inria.fr/gitlab/>](https://app-learninglab.inria.fr/gitlab/)
Si vous accédez à ce lien depuis la plate-forme FUN, vous n'avez pas à vous authentifier et vous pouvez facilement lire et modifier tous vos fichiers. C'est très pratique, mais dans la plupart des cas, vous voudrez avoir votre propre copie locale du référentiel et vous devrez synchroniser votre copie locale avec celle de GitLab. Vous devrez évidemment vous authentifier sur GitLab pour propager vos modifications.
Ce document décrit le logiciel que vous devez installer sur votre ordinateur et comment gérer l'authentification. La section "Configuration de Git" est illustrée dans un [tutoriel vidéo](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session01bis/jump_to_id/7508aece244548349424dfd61ee3ba85)(en français).
Veuillez lire attentivement toutes ces instructions, en particulier celle sur la "Configuration de votre mot de passe sur GitLab".
Table des matières<span class="tag" data-tag-name="TOC"></span>
-[Dire à Git qui vous êtes : nom et email](#dire-à-git-qui-vous-êtes-nom-et-email)
-[Gérer les proxy](#gérer-les-proxy)
-[Récupérer votre mot de passe par défaut sur GitLab (et le changer éventuellement)](#récupérer-votre-mot-de-passe-par-défaut-sur-gitlab-et-le-changer-éventuellement)
-[Enregistrer votre mot de passe localement](#enregistrer-votre-mot-de-passe-localement)
-[Optionnel : authentification par SSH](#optionnel-authentification-par-ssh)
-[Utiliser Git par lignes de commandes pour synchroniser vos fichiers locaux avec Gitlab](#utiliser-git-par-lignes-de-commandes-pour-synchroniser-vos-fichiers-locaux-avec-gitlab)
Installer Git
=============
Linux (Debian, Ubuntu)
----------------------
Nous ne fournissons ici que des instructions pour les distributions basées sur Debian. N'hésitez pas à contribuer à ce document en fournissant des informations à jour sur les autres distributions (RedHat, Fedora, par exemple).
Run (as root) :
``` bash
apt-get update ; apt-get install git
```
Mac OSX et Windows
------------------
- Télécharger et installer Git depuis le [site Git](https://git-scm.com/downloads).
- Clients Git optionnels (ne devraient pas être nécessaires si vous travaillez dans RStudio) :
-[SourceTree](https://www.sourcetreeapp.com/)
-[GitHub Desktop](https://desktop.github.com/)
> Semble fonctionner avec GitLab et https (voir [discussion](https://github.com/desktop/desktop/issues/852)).
Configurer Git
==============
Dire à Git qui vous êtes : nom et email
---------------------------------------
1. Ouvrir un terminal.
2. Définir un nom d'utilisateur et un email dans Git :
3. Confirmer que vous avez correctement défini le nom d'utilisateur et l'email Git :
``` shell
git config --global user.name
git config --global user.email
```
``` example
Mona Lisa
email@example.com
```
Gérer les proxy
---------------
Vous êtes peut-être derrière un proxy, auquel cas vous pouvez avoir des problèmes de clonage ou d’extraction à partir d’un dépôt distant, ou une erreur telle que : `unable to access... Couldn't resolve host...`
Le `proxyPassword` sera stocké en texte brut (non crypté) dans votre fichier `.gitconfig`, ce que vous ne souhaitez peut-être pas. Dans ce cas, supprimez-le de l'URL et vous serez invité à le saisir chaque fois que vous en aurez besoin.
Récupérer votre mot de passe par défaut sur GitLab (et le changer éventuellement)
**Avertissement (utilisateurs Jupyter) :** changer votre mot de passe Gitlab par défaut vous empêchera de commiter les notebooks Jupyter que nous avons déployés pour le MOOC. Vous devrez effectuer l’étape supplémentaire de modification de votre `~/.git-credentials` dans l’environnement Jupyter (éventuellement plusieurs fois).
1. Récupérer votre mot de passe par défaut en utilisant l'outil [Gitlab credentials retrieval](https://app-learninglab.inria.fr/jupyterhub/services/password) comme décrit dans la [ressource correspondante](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session01bis/jump_to_id/7508aece244548349424dfd61ee3ba85).

La première séquence de caractères longue et laide est votre identifiant GitLab qu'il est facile de trouver une fois que vous êtes connecté à Gitlab. Cependant la seconde séquence de caractères est votre mot de passe et cette page Web est le seul endroit où vous pouvez le trouver. Nous avons utilisé le mécanisme d'authentification FUN pour propager vos informations d'identification de sorte que seulement vous puissiez y accéder. Vous devrez utiliser ce mot de passe lorsque vous essaierez de propager des modifications de votre ordinateur vers GitLab.
*Note : Vous devez accéder à cette page Web à partir de la* *plate-forme FUN, sinon vous risquez d'obtenir une erreur 405 en* *essayant d'accéder directement à <https://app-learninglab.inria.fr/jupyterhub/services/password>.*

2. Accéder à [GitLab](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session01bis/xblock/block-v1:inria+41016+session01bis+type@lti+block@05a0ce425f1741e5bee5049040f70529/handler/preview_handler).
*Note : Vous devez à nouveau accéder à Gitlab à partir de la plate-forme* *FUN, sinon vous risquez d'obtenir une erreur 405 en essayant d'accéder directement à* <https://app-learninglab.inria.fr/gitlab/users/sign_in/>.
3. Cliquez sur le premier bouton `Sign in`. Vous pouvez également utiliser l'identifiant/mot de passe que vous venez de récupérer et utiliser le second bouton `Sign in`. Le second bouton permet de plus de se connecter sans passer par la plateforme FUN une fois que vous connaissez votre identifiant/mot de passe.

4. Si vous souhaitez modifier votre mot de passe, accédez à `Account >
Settings > Password` et définissez votre mot de passe à l'aide du mot de passe par défaut que vous venez de récupérer. Encore une fois, si vous utilisez les notebooks Jupyter que nous avons déployés pour le MOOC, n’oubliez pas que changer votre mot de passe Gitlab par défaut vous empêchera de les commiter. Vous devrez effectuer l’étape supplémentaire consistant à changer votre `~/.git-credentials` Jupyter via une console Jupyter (voir section suivante).

Enregistrer votre mot de passe localement
-----------------------------------------
Si vous clonez votre dépôt en collant simplement l'URL de GitLab, vous serez invité à saisir votre identifiant et votre mot de passe chaque fois que vous souhaitez propager vos modifications locales, ce qui est fastidieux. C’est pourquoi vous pouvez demander à Git de se rappeler de votre identifiant et votre mot de passe comme suit :
``` shell
git config --global credential.helper cache # remember my password
git config --global credential.helper "cache --timeout=3600"# for one hour at most
```
Avec cette configuration, vous serez invité à saisir votre mot de passe, mais celui-ci sera mis en cache et ne sera plus demandé pendant une heure. Vous voudrez peut-être lire [ces instructions](https://stackoverflow.com/questions/5343068/is-there-a-way-to-skip-password-typing-when-using-https-on-github) pour mieux comprendre comment tout cela fonctionne.
Si vous souhaitez que votre mot de passe soit mémorisé en permanence, vous devez utiliser cette commande :
``` shell
git config credential.helper store
```
Votre mot de passe sera alors stocké dans un fichier `.git-credentials` en texte brut (non-crypté). Sur une machine parfaitement sécurisée, cela peut être très bien... ou pas... ;) Utilisez cette possibilité à vos risques et périls.
Optionnel : authentification par SSH
------------------------------------
Il existe deux manières d'authentifier et de synchroniser votre dépôt local avec GitLab : via HTTPS ou via SSH. Le premier est ce qui vient d'être décrit et ne nécessite aucune installation de logiciel particulier sur votre machine, c'est donc ce que je recommande pour ce MOOC. Pourtant, je préfère le second (bien que cela puisse paraître un peu plus technique), c'est pourquoi je le décris ici. Cela consiste à installer SSH, à créer une paire de clés ou des clés privée/publique et à télécharger votre clé publique SSH sur GitLab. Cette section fournit des informations sur la procédure à suivre.
### Installer SSH
1. Linux (Debian, Ubuntu)
Nous ne fournissons ici que des instructions pour les distributions basées sur Debian. N'hésitez pas à contribuer à ce document en fournissant des informations à jour sur les autres distributions (RedHat, Fedora, par exemple).
Run (as root) :
``` bash
apt-get update ; apt-get install openssh-client
```
2. macOS
C'est installé par défaut donc vous n'avez rien à faire.
3. Windows
Vous devez installer le client [Putty](https://www.ssh.com/ssh/putty/windows/). Une fois l’installation terminée, suivez la section [PuTTYgen - Key Generator for PuTTY on Windows](https://www.ssh.com/ssh/putty/windows/puttygen).
### Configurer SSH sur GitLab
Vous trouverez [ici](https://docs.gitlab.com/ee/ssh/) les explications officielles sur la configuration de votre clé SSH sur GitLab. Vous pouvez aussi regarder cette vidéo :
Cette section décrit un moyen générique (par lignes de commandes) de synchroniser vos fichiers locaux avec Gitlab. Vous n'en aurez pas besoin si vous suivez le parcours Jupyter. Si vous suivez le parcours RStudio, toutes ces opérations peuvent être effectuées via RStudio et vous voudrez peut-être lire [les instructions correspondantes](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session01bis/jump_to_id/1a4f58a1efed437c93a9f5c5f15df428). Si vous suivez le chemin Org-Mode, toutes ces opérations peuvent être effectuées via Magit et vous voudrez peut-être lire [les instructions correspondantes](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session01bis/jump_to_id/508299f7373449a3939faa5b11462bc4).
Voici d'autres moyens d'apprendre Git par lignes de commandes :
- Le [Software Carpentry git tutorial](http://swcarpentry.github.io/git-novice/)
- Le livre Pro Git (gratuit) en [englais](https://git-scm.com/book/en/v2) ou en [français](https://git-scm.com/book/fr/v2). Les deux premiers chapitres suffisent pour bien commencer.
- Le site [Apprenez Git Branching](https://learngitbranching.js.org/) permet d'apprendre Git interactivement et de comprendre les branches.
Maintenant, commençons !
1. Récupérer l'URL du dépôt

2. Cloner le dépôt
``` shell
cd /the/directory/where/you/want/to/clone/your/repository
Alternativement, vous pouvez vouloir indiquer votre identifiant maintenant, bien que je vous suggère plutôt de suivre les instructions de la partie *Enregistrer votre mot de passe localement*.
Maintenant un répertoire `mooc-rr` a été créé sur votre ordinateur.
3. Inspecter le répertoire correspondant au dépôt
``` shell
cd mooc-rr
ls # (Unix)
dir # (Windows)
```
4. Synchroniser avec GitLab
Vous devez indiquer les fichiers à suivre (`git add`) et les valider localement (`git commit`) avant de pouvoir les transférer (`git push`) à GitLab. Le `git status` vous indiquera si les fichiers sont suivis/modifiés/commités/...
Supposons que vous venez de créer un fichier `fichier.txt` à la racine du répertoire `mooc-rr`.
``` shell
git status
```

``` shell
git add fichier.txt
git status
```

``` shell
git commit -m "message commit"
```

``` shell
git status
```

Le fichier peut ensuite être transféré vers GitLab :
``` shell
git push
```
À ce stade, Git vous demandera votre identifiant/mot de passe, sauf si vous avez suivi les instructions de la partie *Enregistrer votre mot de passe localement*.
NB : vous ne serez pas autorisé à propager vos modifications dans GitLab si d'autres modifications ont été propagées entre temps (par exemple par quelqu'un d'autre).

5. Synchronisation à partir de Gitlab : pour éviter le problème précédent, vous devez d’abord récupérer les modifications distantes de GitLab et les appliquer localement.
``` shell
git pull
```
Alors seulement pourrez-vous exécuter le `git push`.
-[Creating or importing a notebook](#creating-or-importing-a-notebook)
lists several Jupyter tricks (in particular, it illustrates many
-[Running R and Python in the same notebook](#running-r-and-python-in-the-same-notebook)
`IPython magic` commands) that should improve your efficiency (note that
-[Other languages](#other-languages)
this blog post is about two years old so some of the tricks may have
-[2. Installing and configuring Jupyter on your computer](#2-installing-and-configuring-jupyter-on-your-computer)
been integrated in the default behavior of Jupyter now).
-[2.1 Installing Jupyter](#21-installing-jupyter)
-[2.2 Making sure Jupyter allows you to use R](#22-making-sure-jupyter-allows-you-to-use-r)
-[2.3 Additional tips](#23-additional-tips)
## Creating or importing a notebook
1. Jupyter tips and tricks
==========================
Using the Jupyter environment we deployed for this MOOC will allow to
The following [webpage](https://www.dataquest.io/blog/jupyter-notebook-tips-tricks-shortcuts/) lists several Jupyter tricks (in particular, it illustrates many `IPython magic` commands) that should improve your efficiency (note that this blog post is about two years old so some of the tricks may have been integrated in the default behavior of Jupyter now).
easily access any file from your default GitLab project. There are
situations however where you may want to play with other notebooks.
- Adding a brand new notebook in a given directory
Creating or importing a notebook
Simply follow the following steps:
--------------------------------
1. From the menu: `File -> Open`. You're now in the Jupyter file
Using the Jupyter environment we deployed for this MOOC will allow to easily access any file from your default GitLab project. There are situations however where you may want to play with other notebooks.
manager.
2. Navigate to the directory where you want your notebook to be
Adding a brand new notebook in a given directory
created.
Simply follow the following steps:
3. Then from the top right button: `New -> Notebook: Python 3`.
1. From the menu: `File -> Open`. You're now in the Jupyter file manager.
2. Navigate to the directory where you want your notebook to be created.
4. Give your notebook a name from the menu: `File -> Rename`.
3. Then from the top right button: `New -> Notebook: Python 3`.
4. Give your notebook a name from the menu: `File -> Rename`.
N.B.: If you create a file by doing `File -> New Notebook ->
N.B.: If you create a file by doing `File -> New Notebook ->
Python 3`, the new notebook will be created in the current
Python 3`, the new notebook will be created in the current directory. Moving it afterward is possible but a bit cumbersome (you'll have to go through the Jupyter file manager by following the menu `File -> Open`, then select it, `Shut` it `down`, and `Move` and/or `Rename`).
directory. Moving it afterward is possible but a bit cumbersome
(you'll have to go through the Jupyter file manager by following
Importing an already existing notebook
the menu `File -> Open`, then select it, `Shut` it `down`, and
If your notebook is already in your GitLab project, then simply synchronize by using the `Git pull` button and use the `File -> Open` menu. Otherwise, imagine, you want to import the [following notebook](https://app-learninglab.inria.fr/gitlab/moocrr-session1/moocrr-reproducibility-study/blob/master/src/Python3/challenger.ipynb) from someone else's repository to re-execute it.
`Move` and/or `Rename`).
1. Download the file on your computer. E.g., for this [GitLab hosted notebook](https://app-learninglab.inria.fr/gitlab/moocrr-session1/moocrr-reproducibility-study/blob/master/src/Python3/challenger.ipynb), click on `Open raw` (a small `</>` within a document icon) and save (`Ctrl-S` on most browsers) the content (a long Json text file).
- Importing an already existing notebook
2. Open the Jupyter file manager from the menu `File -> Open` and navigate to the directory where you want to upload your notebook.
If your notebook is already in your GitLab project, then simply
3. Then from the top right button, `Upload` the previously downloaded notebook and confirm the upload.
synchronize by using the `Git pull` button and use the `File ->
4. Open the freshly uploaded notebook through the Jupyter file manager.
Open` menu. Otherwise, imagine, you want to import the [following
Note that this `%%R` notation indicates that R should be used for the whole cell but an other possibility is to use `%R` to have a single line of R within a python cell.
Jupyter is not limited to Pytyhon and R. Many other languages are available: [<https://github.com/jupyter/jupyter/wiki/Jupyter-kernels>](https://github.com/jupyter/jupyter/wiki/Jupyter-kernels), including non-free languages like SAS, Mathematica, Matlab... Note that the maturity of these kernels differs widely.
None of these other languages have been deployed in the context of our MOOC but you may want to read the next sections to learn how to set up your own Jupyter on your computer and benefit from these extensions.
Since the question was asked several times, if you really need to stay with SAS, you should know that SAS can be used within Jupyter using either the [Python SASKernel](https://sassoftware.github.io/sas_kernel/) or the [Python SASPy](https://sassoftware.github.io/saspy/) package (step by step explanations about this are given [here](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/documents/tuto_jupyter_windows/tuto_jupyter_windows.md)).
Since proprietary software such as SAS cannot easily be inspected, we discourage its use as it hinders reproducibility by essence. But perfection does not exist anyway and using Jupyter literate programming approach allied with systematic control version and environment control will certainly help anyway.
Note that Jupyter notebooks are only a small part of the picture and that Jupyter is now part of a bigger project: [JupyterLab](https://blog.jupyter.org/jupyterlab-is-ready-for-users-5a6f039b8906), which allows you to mix various components (including notebooks) in your browser. In the context of this MOOC, our time frame was too short to benefit from JupyterLab which was still under active development. You may, however, prefer JupyterLab when doing an installation on your own computer.
Follow these instructions if you wish to have a Jupyter environment on your own computer similar to the one we set up for this MOOC.
yourowncomputersimilartotheonewesetupforthisMOOC.
First,downloadandinstallthe[latestversionof
First, download and install the [latest version of Miniconda](https://conda.io/miniconda.html). We use Miniconda version `4.5.4` and Python version `3.6` on our server.
Miniconda is a light version of Anaconda, which includes Python, the Jupyter Notebook, and other commonly used packages for scientific computing and data science.
Then download the [mooc<sub>rr</sub> environment file](https://gist.github.com/brospars/4671d9013f0d99e1c961482dab533c57) and create the environment using conda:
The environment described in the last section should include R, but if
The environment described in the last section should include R, but if you proceeded otherwise and only have Python available in Jupyter, you may want to read the following section.
you proceeded otherwise and only have Python available in Jupyter, you
Here is what we had to install on a recent Debian computer to make sure
Here is what we had to install on a recent Debian computer to make sure the notebook export via LaTeX works:
the notebook export via LaTeX works:
``` shell
``` shell
sudo apt-get install texlive-xetex wkhtmltopdf
sudo apt-get install texlive-xetex wkhtmltopdf
```
```
Obviously, you can convert to html or pdf using the using the `File >
Obviously, you can convert to html or pdf using the using the `File > Download as > HTML` (or `PDF`) menu option. This can also be done from the command line with the following command:
Download as > HTML` (or `PDF`) menu option. This can also be done from
the command line with the following command:
``` bash
``` bash
ipython3 nbconvert --to pdf Untitled.ipynb
ipython3 nbconvert --to pdf Untitled.ipynb
```
```
If you want to use a specific style, then the `nbconvert` exporter
If you want to use a specific style, then the `nbconvert` exporter should be customized. This is discussed and demoed [here](http://markus-beuckelmann.de/blog/customizing-nbconvert-pdf.html). We encourage you to simply read the [doc of nbconvert](https://nbconvert.readthedocs.io/en/latest/).
should be customized. This is discussed and demoed
Instead of going directly through LaTeX and playing too much with the
Instead of going directly through LaTeX and playing too much with the `nbconvert` exporter, an other option consists in exporting to Markdown and playing with [pandoc](https://pandoc.org/). Both approaches work, it's rather a matter of taste.
`nbconvert` exporter, an other option consists in exporting to Markdown
and playing with [pandoc](https://pandoc.org/). Both approaches work,
it's rather a matter of taste.
**Windows**
**Windows**
Download and install MiKTeX from the [MiKTeX
Download and install MiKTeX from the [MiKTeX webpage](https://miktex.org/download) by choosing the right operating system. You will be prompted to install some specific packages when exporting to pdf.
webpage](https://miktex.org/download) by choosing the right operating
system. You will be prompted to install some specific packages when
exporting to pdf.
### • Improving notebook readability
### • Improving notebook readability
Here are a few extensions that can ease your life:
Here are a few extensions that can ease your life:
- [Code
-[Code folding](https://stackoverflow.com/questions/33159518/collapse-cell-in-jupyter-notebook) to improve readability when browsing the notebook.
to improve readability when browsing the notebook.
``` shell
``` shell
pip3 install jupyter_contrib_nbextensions
pip3 install jupyter_contrib_nbextensions
# jupyter contrib nbextension install --user # not done yet
# jupyter contrib nbextension install --user # not done yet
```
```
- [Hiding code](https://github.com/kirbs-/hide_code) to improve
-[Hiding code](https://github.com/kirbs-/hide_code) to improve readability when exporting.
readability when exporting.
``` bash
``` bash
sudo pip3 install hide_code
sudo pip3 install hide_code
...
@@ -278,28 +214,8 @@ Here are a few extensions that can ease your life:
...
@@ -278,28 +214,8 @@ Here are a few extensions that can ease your life:
### • Interacting with GitLab and GitHub
### • Interacting with GitLab and GitHub
To ease your experience, we added pull/push buttons that allow you to
To ease your experience, we added pull/push buttons that allow you to commit and sync with GitLab. This development was specific to the MOOC but inspired from a previous [proof of concept](https://github.com/Lab41/sunny-side-up). We have recently discovered that someone else developed about at the same time a [rather generic version of this Jupyter plugin](https://github.com/sat28/githubcommit). Otherwise, remember that it is very easy to insert a shell cell in Jupyter in which you can easily issue git commands. This is how we work most of the time.
commit and sync with GitLab. This development was specific to the MOOC
but inspired from a previous [proof of
This being said, you may have noticed that Jupyter keeps a perfect track of the sequence in which cells have been run by updating the "output index". This is a very good property from the reproducibility point of view but depending on your usage, you may find it a bit painful when committing. Some people have thus developed [specific git hooks](https://gist.github.com/pbugnion/ea2797393033b54674af) to ignore these numbers when committing Jupyter notebooks. There is a long an interesting discussion about various options on [StackOverflow](https://stackoverflow.com/questions/18734739/using-ipython-notebooks-under-version-control).
concept](https://github.com/Lab41/sunny-side-up). We have recently
discovered that someone else developed about at the same time a [rather
For those who use [JupyterLab](https://blog.jupyter.org/jupyterlab-is-ready-for-users-5a6f039b8906) rather than the plain Jupyter, a specific [JupyterLab git plugin](https://github.com/jupyterlab/jupyterlab-git) has been developed to offer a nice version control experience.
generic version of this Jupyter
plugin](https://github.com/sat28/githubcommit). Otherwise, remember that
it is very easy to insert a shell cell in Jupyter in which you can
easily issue git commands. This is how we work most of the time.
This being said, you may have noticed that Jupyter keeps a perfect track
of the sequence in which cells have been run by updating the "output
index". This is a very good property from the reproducibility point of
view but depending on your usage, you may find it a bit painful when
committing. Some people have thus developed [specific git
hooks](https://gist.github.com/pbugnion/ea2797393033b54674af) to ignore
these numbers when committing Jupyter notebooks. There is a long an
Since a few years, we systematically require any or our students to have a laboratory notebook in org-mode. Most of the time, they start in private repositories but often end up being fully opened. Here are a few ones:
- Luka Stanisic (a former PhD student advised by Arnaud Legrand) starting using this methodology during his Msc and developed further throughout his PhD. Part of his [PhD thesis](http://mescal.imag.fr/membres/luka.stanisic/thesis/thesis.pdf) was actually about designing a methodology for reproducible experiments in large scale distributed systems. You may want to have a look at [his postdoc LabBook](http://starpu-simgrid.gforge.inria.fr/) and to the [report of Léo Villeveygoux](https://framagit.org/lvgx/pfe/blob/master/doc/labbook.org) whom he advised.
- Tom Cornebize is currently a PhD student advised by Arnaud Legrand and during his MSc, he also heavily [loged his activity on Github](https://github.com/Ezibenroc/simulating_mpi_applications_at_scale).
-[Lucas Schnorr](https://github.com/schnorr)'s students usually also maintain their journal in a very nice way: [Tais Bellini's BSc.](https://github.com/taisbellini/aiyra/blob/master/LabBook.org), [Arthur Krause's LabBook](https://github.com/mittmann/hpc/blob/master/LabBook.org), [Luca Nesi's LabBook](http://www.inf.ufrgs.br/~llnesi/memory_report/MemoryReport.html).
-[Martin Quinson](https://people.irisa.fr/Martin.Quinson/Research/Students/Methodo/)'s students also follow such conventions:
- Ezequiel Torti Lopez, M2R 2014. [Report](https://github.com/mquinson/simgrid-simpar/blob/master/report.org), with both the data provenance and the data analysis included in the appendix.
- Gabriel Corona, engineer on SimGrid, 2015-2016. [Journal](https://github.com/randomstuff/simgrid-journal/blob/master/journal.org), [Blog (findings)](http://www.gabriel.urdhr.fr/tags/simgrid/).
- Matthieu Nicolas, engineer on PLM, 2014-2016, [Journal](https://github.com/MatthieuNICOLAS/PLM-reporting/blob/master/activity-report.org).
Org-mode is obviously not the only option and many of our students use am mixture of org-mode, rstudio and jupyter depending on what is more convenient.
How to report efficiently (by Martin Quinson)
=============================================
My friend Martin has gathered \[\[<https://people.irisa.fr/Martin.Quinson/Research/Students/Methodo/>\]\[an excellent compendium of information and references on his webpage to explain his students what he expects from them\]\]. **I'll therefore simply paraphrase him here** with the most important aspects related to reporting but feel free to read [the original version](https://people.irisa.fr/Martin.Quinson/Research/Students/Methodo/):
Reporting
---------
I ask you to write a little reporting regularly. Depending on the situation, it may be every day, every week or every month. In any case, your reporting is very important for the following reasons:
- It forces you to think about what you are doing, which may help you to unblock your problem by your own. Writing down the problems in a clear way is often sufficient to see the solution appearing.
- It helps me following your progress even between the meetings. I cannot unblock you if I don't detect that you are on a wrong lead or otherwise blocked.
- It keeps a track of the steps in your work. That's good for the day where you want to write your final report (even if a final report should never be presented in the chronological order). That's good for the next after you who will be supposed to continue you effort, or to build upon it.
- That person may be yourself (if you go for a PhD program), another intern, myself or even someone else on the Internet: that's what we call Open Science, an effort where everyone can build upon the scientific work of everyone.
I want you to write your reporting in an org file (yep, you don't have a choice here). \[..\]
Reporting Logistics
-------------------
Once you're setup with all software installed and somehow configured, you need to create a reporting file in a place where I can see it and where it won't get lost if your disk crashes or something. Open a dedicated git repository (on github, gitorious, gitlab, ...) for that. After your internship, your report should be archived directly in the source tree of the software that you are working on, if any. But having your reporting located in the source tree may complicate things during your work.
Yes, it means that your file will be public at some point, but that's why we call it "Open Science", after all. Also, you should write it in English if possible. The part of your reporting that is called "Journal" (see below) may be written in French if you are more efficient this way but the rest must be in English. Don't make your tone too formal because the file is public. Make it efficient. Nobody will ever blame you for the work you did during an internship a long time ago. If you really want, we can even make this file anonymous. Just speak to me.
You want to write your reporting before leaving work. Weekly reporting should be written on Friday, one or two hours before leaving. That's the best solution to have a nice week end without thinking about work, and still lose no information that you would need on Monday morning.
Reporting Document Organization
-------------------------------
Your reporting document should have four main parts:
Findings
This section summarizes the general information that you gathered during your work. It is empty at the beginning of your internship, and gets fleshed with the important things that you find on your way. That's where bibliographical information go, for example. But that's definitely not where TODO notes go (see below).
Development
This section presents the technical sides of your work. Don't write anything in there yet. Put it all in the Journal part for now.
Journal
Describe the day-to-day work done for each period (day, week or month) of your internship. That's the most important part of your reporting, and we come back to it below.
Conclusion
That's what you write in the next week of your internship. You can see it as a letter to the next guy, explaining the current state of your work, a few words about its technical organization, and what should be done next on that topic. Keep this part highly technical, the overall organization of your internship will be seen in your final report.
The Journal part is the only part that you may write in French on need. You want to add one subsection per period to your journal. Don't make it too long, or you would waste time writing long texts that very few will ever read. Don't make it too short or it will be impossible to understand it on Monday morning (or three months after). Finding the good balance is sometimes difficult, but I will provide feedback on your first entries, so don't worry.
Each of section describing a period should contain three subsubsections:
Things done
a few words about what you've done. Something like 2 or 4 items with a few words describing what you've done. You can omit the title of that section and put the items directly in the upper section (see the example below).
Blocking points and questions
try to explain clearly the things that block you or slow you down. If you found the solution already, then it should be part of the previous subsection (but you should say a few words nevertheless). Also ask every question that you may have for me in that section. If the question are personal (e.g., about the logistics of your internship such as salary or so), please prefer emails that are not publicly visible. If this section is empty for a given period, skip it all together (no empty subsubsections).
Planned work
A few items about what you plan to work on during the next period.
A template of reporting file is given at the end of this section. This is just a strong advice: If you really feel better with another file organization, then give it a try for one period, and ask for my feedback. I can adapt, and I do not pretend that my advice is the definitive answer. It's just the result of my experience so far.
Notice how TODO items are written: they are given as items in the Planned work sections of the journal. As explained in the [documentation](http://orgmode.org/manual/Checkboxes.html), you simply have to write "\[\]" in front of items that you plan to do in the future.
You should add a \[1/\] on the "Planned work" line, so that emacs keeps track of what is done and what is still to do. Once they are done, you type C-c C-C on their lines to change the blank box \[\] into a checked box \[X\]. Also, the \[1/\] will be changed to denote the amount of work that is still to be done.
At any point, you can see all ongoing TODO items with the following keystrokes: "C-c / t". More information on TODOs in orgmode's [documentation](http://orgmode.org/manual/TODO-basics.html). The important thing here is that most TODO items must only be written in the *Journal* part (so that we know when they occurred).
**Do not edit past entries of your journal**, unless you have very good reasons. If you must, make sure that you don't lose information about the path that you took (remember the Open Science thingy). You should always **add** information to past entries, such as:
``` shell
- *edit* This hypothesis does not hold; see the entry of [the day where you found it] for more information.
```
The only exception are TODO entries, that should clearly be rewritten to DONE entries. If you need to adapt your TODO entry (because the initial goal was poorly stated or otherwise), change the initial entry from TODO to CANCELED (or check the box after stating in a subitem that it was not done but canceled, and why), and create a new TODO entry in the current period section.
``` example
* Introduction
This file contains the reporting for my beloved internship done on
this topic on that year. For now, just add the official title of
your internship (check the convention signed between your
university and my lab). After a few weeks, once you really
understand your internship, you should write a few paragraphs about
the context, problem and motivation of your work, with some
-[Using Git from RStudio](#using-git-from-rstudio)
-[Cloning a repository](#cloning-a-repository)
-[Modifying a file](#modifying-a-file)
Installing RStudio
==================
Linux (debian, ubuntu)
----------------------
We provide here only instructions for Debian-based distributions. Feel free to contribute to this document to provide up-to-date information for other distributions (e.g., RedHat, Fedora).
Today, the stable versions of the most common distributions provide recent enough versions of R:
- Debian (stretch) ships with [R 3.3.3-1](https://packages.debian.org/stretch/r-base), [knitr 1.15.1](https://packages.debian.org/stretch/r-cran-knitr), and [ggplot 2.2.1](https://packages.debian.org/stretch/r-cran-ggplot2)
- Ubuntu (bionic 18.04) ships with [R 3.4.4](https://packages.ubuntu.com/bionic/r-base), and [knitr 1.17](https://packages.ubuntu.com/bionic/r-cran-knitr), and [ggplot 2.2.1](https://packages.ubuntu.com/bionic/r-cran-ggplot2)
- Ubuntu (artful 17.04) ships with [R 3.4.2](https://packages.ubuntu.com/artful/r-base), and [knitr 1.15](https://packages.ubuntu.com/artful/r-cran-knitr), and [ggplot 2.2.1](https://packages.ubuntu.com/artful/r-cran-ggplot2)
If your distribution is older than this, well, it may be a good time for upgrading...
### Installing R
First, you need to install the R language and convenient packages by running (as root):
Alternatively, if the installation of `r-cran-gplot2` or `r-cran-knitr` fails, you may want to install them locally (through the R packaging system) and manually by running the following commands in R (or RStudio):
``` r
install.packages("knitr")
install.packages("ggplot2")
```
If you plan to export pdf documents with LaTeX, you probably also want to run (as root):
``` bash
apt-get update ; apt-get install texlive-base
```
### Installing RStudio
RStudio is unfortunately not packaged within Debian so the easiest is to download the corresponding Debian package on the [RStudio webpage](https://www.rstudio.com/products/rstudio/download/#download) and then to install it manually (you may have to adjust the version number in the following example). Here is how to install it:
> Some instructions on installing R and knitr must be missing. This should be tested and improved.
- Download and install R from the [CRAN webpage](https://cran.r-project.org/) by choosing the right operating system.
- Download and install RStudio from the [RStudio webpage](https://www.rstudio.com/products/rstudio/download/#download) by choosing the right operating system.
- Download and install MiKTeX from the [MiKTeX webpage](https://miktex.org/download) by choosing the right operating system. You will be prompted to install some specific packages when exporting to pdf.
- Open RStudio and type the following commands in the console to install `knitr` and `ggplot2`:
``` r
install.packages("knitr",dep=TRUE)
install.packages("ggplot2",dep=TRUE)
```
RStudio documentation
=====================
The RStudio team has created a lot of very good material and tutorials. You should definitively look at the [Cheat sheets webpage](https://www.rstudio.com/resources/cheatsheets/). In particular you may want to have look at the following ones:
-[R Markdown](https://github.com/rstudio/cheatsheets/raw/master/rmarkdown-2.0.pdf)(here is also a [nice step-by-step presentation of Rmarkdown](https://rmarkdown.rstudio.com/)),
- The [R Markdown Reference guide](https://www.rstudio.com/wp-content/uploads/2015/03/rmarkdown-reference.pdf),
-[Data visualization with ggplot2](https://github.com/rstudio/cheatsheets/raw/master/data-visualization-2.1.pdf),
-[Data transformation with dplyr](https://github.com/rstudio/cheatsheets/raw/master/data-transformation.pdf)
In case it helps, here are some (sometimes outdated) French versions of these documents:
-[Visualisation de données avec ggplot2](https://github.com/rstudio/cheatsheets/raw/master/translations/french/ggplot2-french-cheatsheet.pdf)
-[Transformation de données avec dplyr](https://github.com/rstudio/cheatsheets/raw/master/translations/french/data-wrangling-french.pdf)
-[Un court document sur R Markdown](https://www.fun-mooc.fr/c4x/UPSUD/42001S02/asset/RMarkdown.pdf)
Using Git from RStudio
======================
If you have never used git with RStudio, **we strongly advise that you follow [our tutorial on using git from RStudio](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/d132a854b0464ad29085cedaded23136)** (/"RStudio et Gitlab"/ in French). Before proceeding, make sure you also have followed the **["git/GitLab configuration" tutorial](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/7508aece244548349424dfd61ee3ba85)** (in French).
Alternatively, you may want to watch [this video](https://www.youtube.com/embed/uHYcDQDbMY8)(in English). If you do not like videos, you should have a look at the [step-by-step explanations from Software Carpentry](https://swcarpentry.github.io/git-novice/14-supplemental-rstudio/index.html). It comes with many screenshots and is quite progressive.
Cloning a repository
--------------------
Open RStudio and do the following steps:
- Create a new version controled project: `File / New Project / Version Control`


- Get the URL from your GitLab repository:

- Indicate this URL in the "Repository URL" field (*you may want to prefix this URL with `xxx@` where `xxx` is**your Gitlab id to avoid repeatedly giving it later on*).

- If you're behind a proxy, git should be configured accordingly. Check the ["Dealing with proxies" section](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/7508aece244548349424dfd61ee3ba85).
- Git will then connect to Gitlab and fetch a whole copy of the repository.
- RStudio should restart in a mode related to Git:

- The file manager on the right, allows you to browse the version controled repository.
Modifying a file
----------------
- Open `Module2/exo1/toy_document.Rmd` and perform a simple modification.
- Save
- Go to the Git menu to commit


- Select the lines to commit and then click on `commit`

Your modifications have now been commited on your local machine. They haven't been propagated to GitLab yet.
- Click on `push` to propagate them on GitLab



**NB**: You won't be able to propagate your modifications on GitLab if some modifications have been done on GitLab in the meantime. 
- You should first merge these remote modifications locally. Click on `pull` to get these modifications on your machine.
-[Utiliser Git avec RStudio](#utiliser-git-avec-rstudio)
-[Cloner un dépôt](#cloner-un-dépôt)
-[Modifier un fichier](#modifier-un-fichier)
Installer RStudio
=================
Linux (debian, ubuntu)
----------------------
Nous ne fournissons ici que des instructions pour les distributions basées sur Debian. N’hésitez pas à contribuer à ce document en fournissant des informations à jour sur les autres distributions (RedHat, Fedora, par exemple).
Aujourd'hui, les versions stables des distributions les plus courantes fournissent des versions assez récentes de R :
- Debian (stretch) est livré avec [R 3.3.3-1](https://packages.debian.org/stretch/r-base), [knitr 1.15.1](https://packages.debian.org/stretch/r-cran-knitr), et [ggplot 2.2.1](https://packages.debian.org/stretch/r-cran-ggplot2)
- Ubuntu (bionic 18.04) est livré avec [R 3.4.4](https://packages.ubuntu.com/bionic/r-base), [knitr 1.17](https://packages.ubuntu.com/bionic/r-cran-knitr), et [ggplot 2.2.1](https://packages.ubuntu.com/bionic/r-cran-ggplot2)
- Ubuntu (artful 17.04) est livré avec [R 3.4.2](https://packages.ubuntu.com/artful/r-base), [knitr 1.15](https://packages.ubuntu.com/artful/r-cran-knitr), et [ggplot 2.2.1](https://packages.ubuntu.com/artful/r-cran-ggplot2)
Si votre distribution est plus ancienne, c'est peut-être l'occasion de la mettre à jour...
### Installer R
Pour commencer, vous devez installer le langage R et quelques packages en exécutant (à la racine) :
Si l'installation de `r-cran-knitr` ou `r-cran-gplot2` échoue, vous pouvez également installer ces packages manuellement en exécutant les commandes suivantes sous R (ou RStudio) :
``` r
install.packages("knitr")
install.packages("ggplot2")
```
Si vous envisagez d'exporter des documents pdf avec LaTeX, il faudra probablement aussi exécuter (à la racine) :
``` shell
apt-get update ; apt-get install texlive-base
```
### Installer RStudio
RStudio n’est malheureusement pas intégré à Debian. Le plus simple est de télécharger le paquet Debian correspondant sur le [site RStudio](https://www.rstudio.com/products/rstudio/download/#download), puis de l’installer manuellement (vous devrez peut-être adapter le numéro de version) :
- Télécharger et installer R depuis le [site CRAN](https://cran.r-project.org/) en choisissant le bon système d'exploitation.
- Télécharger et installer RStudio depuis le [site RStudio](https://www.rstudio.com/products/rstudio/download/#download) en choisissant le bon système d'exploitation.
- Télécharger et installer MiKTeX depuis le [site MiKTeX](https://miktex.org/download) en choisissant le bon système d'exploitation. Vous serez amené à installer différents packages lors du premier export pdf.
- Ouvrir RStudio et exécuter les commandes suivantes dans la console pour installer `knitr` et `ggplot2`
``` r
install.packages("knitr",dep=TRUE)
install.packages("ggplot2",dep=TRUE)
```
Documentation RStudio
=====================
L’équipe de RStudio a créé différents matériels et tutoriels très bien faits. Nous vous recommandons de consulter les [fiches mémo](https://www.rstudio.com/resources/cheatsheets/). En particulier, vous pourriez être intéressés par celles-ci :
-[R Markdown](https://github.com/rstudio/cheatsheets/raw/master/rmarkdown-2.0.pdf)(here is also a [nice step-by-step presentation of Rmarkdown](https://rmarkdown.rstudio.com/)),
- The [R Markdown Reference guide](https://www.rstudio.com/wp-content/uploads/2015/03/rmarkdown-reference.pdf),
-[Data visualization with ggplot2](https://github.com/rstudio/cheatsheets/raw/master/data-visualization-2.1.pdf),
-[Data transformation with dplyr](https://github.com/rstudio/cheatsheets/raw/master/data-transformation.pdf)
Voici aussi les versions françaises de certains documents mais elles ne sont pas toujours à jour :
-[Visualisation de données avec ggplot2](https://github.com/rstudio/cheatsheets/raw/master/translations/french/ggplot2-french-cheatsheet.pdf)
-[Transformation de données avec dplyr](https://github.com/rstudio/cheatsheets/raw/master/translations/french/data-wrangling-french.pdf)
-[Un court document sur R Markdown](https://www.fun-mooc.fr/c4x/UPSUD/42001S02/asset/RMarkdown.pdf)
Utiliser Git avec RStudio
=========================
La première chose à faire est de configurer Git sur votre ordinateur. Pour ce faire, vous pouvez suivre la vidéo [configurer Git pour Gitlab](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/7508aece244548349424dfd61ee3ba85)(en français) et le document [Git et Gitlab](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/module2/ressources/gitlab_fr.org) correspondant (en français).
Vous pourrez alors utiliser Git avec RStudio. Pour ce faire, vous pouvez suivre la vidéo [RStudio - Gitlab](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/d132a854b0464ad29085cedaded23136)(en français) dont les étapes sont reprises ci-dessous.
*(Nous vous signalons aussi cette*[vidéo](https://www.youtube.com/embed/uHYcDQDbMY8)*(en anglais) ainsi que le*[tuto pas à pas](https://swcarpentry.github.io/git-novice/14-supplemental-rstudio/index.html)*(en anglais) de Software Carpentry.)*
Cloner un dépôt
---------------
Ouvrir RStudio et procéder comme suit :
- Créer un nouveau projet sous contrôle de version : `File / New
Project / Version Control`


- Récupérer l'URL du dépôt Gitlab

- Indiquez cette URL dans le champ "Repository URL" *(vous voudrez**peut-être préfixer cette URL avec `xxx@` où `xxx` est votre identifiant**Gitlab pour éviter d'avoir à le ressaisir ultérieurement)*.

- Si vous êtes derrière un proxy, il faut le définir dans Git (voir le paragraphe "Gérer les proxy" de la page sur [Git et Gitlab](https://www.fun-mooc.fr/courses/course-v1:inria+41016+session02/jump_to_id/7508aece244548349424dfd61ee3ba85)).
- Git se connecte à Gitlab et récupère une copie complète du dépôt.
- RStudio redémarre dans un mode lié à Git :

- Le gestionnaire de fichiers à droite vous permet de parcourir le dépôt sous contrôle de version.
Modifier un fichier
-------------------
- Ouvrir le fichier `Module2/exo1/toy_document.Rmd` et le modifier.
- Enregistrer.
- Aller dans le menu Git pour effectuer le commit.


- Sélectionner les lignes à commiter puis cliquer sur `commit`.

Les modifications ont été commitées uniquement sur la machine. Elles n'ont pas été propagées sur Gitlab.
- Cliquer sur `push` pour les propager sur Gitlab.



**NB :** Vous ne pouvez pas propager vos modifications sur GitLab si des modifications ont été faites sur GitLab entre-temps.

- Il faut d’abord récupérer ces modifications distantes sur votre machine locale. Pour ce faire cliquer sur `pull`.
In the MOOC video, I quickly demo how org-mode can be used in various contexts. Here are the (sometimes trimmed) corresponding org-files. These documents depend on many other external data files and are not meant to lead to reproducible documents but it will give you an idea of how it can be organized:
1.[journal.org](journal.org): an excerpt (I've only left a few code samples and links to some resources on R, Stats, ...) from my own journal. This is a personal document where everything (meeting notes, hacking, random thoughts, ...) goes by default. Entries are created with the `C-c c` shortcut.
2.[labbook<sub>single</sub>.org](labbook_single.org): this is an excerpt from the laboratory notebook [Tom Cornebize](https://cornebize.net/) wrote during his Master thesis internship under my supervision. This a personal labbook. I consider this notebook to be excellent and was the ideal level of details for us to communicate without any ambiguity and for him to move forward with confidence.
3.[paper.org](paper.org): this is an ongoing paper based on the previous labbook of Tom Cornebize. As such it is not reproducible as there are hardcoded paths and uncleaned dependencies but writing it from the labbook was super easy as we just had to cut and paste the parts we needed. What may be interesting is the organization and the org tricks to export to the right LaTeX style. As you may notice, in the end of the document, there is a commented section with emacs commands that are automatically executed when opening the file. It is an effective way to depend less on the `.emacs/init.el` which is generally customized by everyone.
4.[labbook<sub>several</sub>.org](labbook_several.org): this is a labbook for a specific project shared by several persons. As a consequence it starts with information about installation, common scripts, has section with notes about all our meetings, a section with information about experiments and an other one about analysis. Entries could have been labeled by who wrote them but there were only a few of us and this information was available in git so we did not bother. In such labbook, it is common to find annotations indicating that such experiment was `:FLAWED:` as it had some issues.
5.[technical<sub>report</sub>.org](technical_report.org): this is a short technical document I wrote after a colleague sent me a PDF describing an experiment he was conducting and asked me about how reproducible I felt it was. It turned out I had to cut and paste the C code from the PDF, then remove all the line numbers and fix syntax, etc. Obviously I got quite different performance results but writing everything in org-mode made it very easy to generate both HTML and PDF and to explicitly explain how the measurements were done.
Here are a few links to other kind of examples:
- Slides: all my slides for a series of lectures is available here: <https://github.com/alegrand/SMPE>. Here is a [typical source](https://raw.githubusercontent.com/alegrand/SMPE/master/lectures/lecture_central_limit_theorem.org) and the [resulting PDF](https://raw.githubusercontent.com/alegrand/SMPE/master/lectures/lecture_central_limit_theorem.pdf)
- Lucas Schnorr, a colleague, maintains:
- a set of templates for various computer science journals/conferences: [IEEE](https://github.com/schnorr/ieeeorg), [Wiley](https://github.com/schnorr/wileyorg), [ACM](https://github.com/schnorr/acmorg), [LNCS](https://github.com/schnorr/llncsorg)
- his lecture on programming languages for undergrads: <https://github.com/schnorr/mlp/tree/master/conteudo>
-[Getting information about your Git repository](#getting-information-about-your-git-repository)
-[Getting information about Python(3) libraries](#getting-information-about-python3-libraries)
-[Getting information about your system](#getting-information-about-your-system)
-[Getting the list of installed packages and their version](#getting-the-list-of-installed-packages-and-their-version)
-[How to list imported modules?](#how-to-list-imported-modules)
-[Saving and restoring an environment with pip](#saving-and-restoring-an-environment-with-pip)
-[Installing a new package or a specific version](#installing-a-new-package-or-a-specific-version)
-[Getting information about R libraries](#getting-information-about-r-libraries)
-[Getting the list imported modules and their version](#getting-the-list-imported-modules-and-their-version)
-[Getting the list of installed packages and their version](#getting-the-list-of-installed-packages-and-their-version-1)
-[Installing a new package or a specific version](#installing-a-new-package-or-a-specific-version-1)
Getting information about your Git repository
=============================================
When taking notes, it may be difficult to remember which version of the code or of a file was used. This is what version control is useful for. Here are a few useful commands that we typically insert at the top of our notebooks in shell cells
``` shell
git log -1
```
``` example
commit 741b0088af5b40588493c23c46d6bab5d0adeb33
Author: Arnaud Legrand <arnaud.legrand@imag.fr>
Date: Tue Sep 4 12:45:43 2018 +0200
Fix a few typos and provide information on jupyter-git plugins.
```
``` shell
git status -u
```
``` example
On branch master
Your branch is ahead of 'origin/master' by 4 commits.
(use "git push" to publish your local commits)
Changes not staged for commit:
(use "git add <file>..." to update what will be committed)
(use "git checkout -- <file>..." to discard changes in working directory)
modified: resources.org
Untracked files:
(use "git add <file>..." to include in what will be committed)
no changes added to commit (use "git add" and/or "git commit -a")
```
*Note: the -u indicates that git should also display the contents of new directories it did not previously know about.*
Then, we often include commands at the end of our notebook indicating how to commit the results (adding the new files, committing with a clear message and pushing). E.g.,
``` shell
git add resources.org;
git commit -m"Completing the section on getting Git information"
git push
```
``` example
[master 514fe2c1 ] Completing the section on getting Git information
To gitlab.inria.fr:learninglab/mooc-rr/mooc-rr-ressources.git
6359f8c..1f8a567 master -> master
```
Obviously, in this case you need to save the notebook before running this cell, hence the output of this final command (with the new git hash) will not be stored in the cell. This is not really a problem and is the price to pay for running git from within the notebook itself.
Getting information about Python(3) libraries
=============================================
Getting information about your system
-------------------------------------
This topic is discussed on [StackOverflow](https://stackoverflow.com/questions/3103178/how-to-get-the-system-info-with-python).
This topic is discussed on [StackOverflow](https://stackoverflow.com/questions/20180543/how-to-check-version-of-python-modules). When using `pip` (the Python package installer) within a shell command, it is easy to query the version of all installed packages (note that on your system, you may have to use either `pip` or `pip3` depending on how it is named and which versions of Python are available on your machine
Here is for example how I get this information on my machine:
``` shell
pip3 freeze
```
``` example
asn1crypto==0.24.0
attrs==17.4.0
bcrypt==3.1.4
beautifulsoup4==4.6.0
bleach==2.1.3
...
pandas==0.22.0
pandocfilters==1.4.2
paramiko==2.4.0
patsy==0.5.0
pexpect==4.2.1
...
traitlets==4.3.2
tzlocal==1.5.1
urllib3==1.22
wcwidth==0.1.7
webencodings==0.5
```
In a Jupyter notebook, this can easily be done by using the `%%sh` magic. Here is for example what you could do and get on the Jupyter notebooks we deployed for the MOOC (note that here, you should simply use the `pip` command):
``` python
%%sh
pipfreeze
```
``` example
alembic==0.9.9
asn1crypto==0.24.0
attrs==18.1.0
Automat==0.0.0
...
numpy==1.13.3
olefile==0.45.1
packaging==17.1
pamela==0.3.0
pandas==0.22.0
...
webencodings==0.5
widgetsnbextension==3.2.1
xlrd==1.1.0
zope.interface==4.5.0
```
In the rest of this document, I will assume the correct command is `pip` and I will not systematically insert the `%%sh` magic.
Once you know which packages are installed, you can easily get additional information about a given package and in particular check whether it was installed "locally" through pip or whether it is installed system-wide. Again, in a shell command:
``` shell
pip show pandas
echo" "
pip show statsmodels
```
``` example
Name: pandas
Version: 0.22.0
Summary: Powerful data structures for data analysis, time series,and statistics
Home-page: http://pandas.pydata.org
Author: None
Author-email: None
License: BSD
Location: /usr/lib/python3/dist-packages
Requires:
Name: statsmodels
Version: 0.9.0
Summary: Statistical computations and models for Python
Without resorting to pip (that will list all available packages), you may want to know which modules are loaded in a Python session as well as their version. Inspired by [StackOverflow](https://stackoverflow.com/questions/4858100/how-to-list-imported-modules), here is a simple function that lists loaded package (that have a `__version__` attribute, which is unfortunately not completely standard).
``` python
defprint_imported_modules():
importsys
forname,valinsorted(sys.modules.items()):
if(hasattr(val,'__version__')):
print(val.__name__,val.__version__)
else:
print(val.__name__,"(unknown version)")
print("**** Package list in the beginning ****");
print_imported_modules()
print("**** Package list after loading pandas ****");
importpandas
print_imported_modules()
```
``` example
**** Package list in the beginning ****
**** Package list after loading pandas ****
_csv 1.0
_ctypes 1.1.0
decimal 1.70
argparse 1.1
csv 1.0
ctypes 1.1.0
cycler 0.10.0
dateutil 2.7.3
decimal 1.70
distutils 3.6.5rc1
ipaddress 1.0
json 2.0.9
logging 0.5.1.2
matplotlib 2.1.1
numpy 1.14.5
numpy.core 1.14.5
numpy.core.multiarray 3.1
numpy.core.umath b'0.4.0'
numpy.lib 1.14.5
numpy.linalg._umath_linalg b'0.1.5'
pandas 0.22.0
_libjson 1.33
platform 1.0.8
pyparsing 2.2.0
pytz 2018.5
re 2.2.1
six 1.11.0
urllib.request 3.6
zlib 1.0
```
Saving and restoring an environment with pip
--------------------------------------------
The easiest way to go is as follows:
``` shell
pip3 freeze > requirements.txt # to obtain the list of packages with their version
pip3 install-r requirements.txt # to install the previous list of packages, possibly on an other machine
```
If you want to have several installed Python environments, you may want to use [Pipenv](https://docs.pipenv.org/). I doubt it allows to track correctly FORTRAN or C dynamic libraries that are wrapped by Python though.
Installing a new package or a specific version
----------------------------------------------
The Jupyter environment we deployed on our servers for the MOOC is based on the version 4.5.4 of Miniconda and Python 3.6. In this environment you should simply use the `pip` command (remember on your machine, you may have to use `pip3`).
If I query the current version of `statsmodels` in a shell command, here is what I will get.
``` shell
pip show statsmodels
```
``` example
Name: statsmodels
Version: 0.8.0
Summary: Statistical computations and models for Python
Home-page: http://www.statsmodels.org/
Author: Skipper Seabold, Josef Perktold
Author-email: pystatsmodels@googlegroups.com
License: BSD License
Location: /opt/conda/lib/python3.6/site-packages
Requires: scipy, patsy, pandas
```
I can then easily upgrade `statsmodels`:
``` shell
pip install--upgrade statsmodels
```
Then the new version should then be:
``` shell
pip show statsmodels
```
``` example
Name: statsmodels
Version: 0.9.0
Summary: Statistical computations and models for Python
Home-page: http://www.statsmodels.org/
Author: Skipper Seabold, Josef Perktold
Author-email: pystatsmodels@googlegroups.com
License: BSD License
Location: /opt/conda/lib/python3.6/site-packages
Requires: scipy, patsy, pandas
```
It is even possible to install a specific (possibly much older) version, e.g.,:
``` shell
pip install statsmodels==0.6.1
```
Getting information about R libraries
=====================================
Getting the list imported modules and their version
The best way seems to be to rely on the `devtools` package (if this package is not installed, you should install it first by running in `R` the command `install.packages("devtools")`).
Some actually advocate that [writing a reproducible research compendium is best done by writing an R package](https://github.com/ropensci/rrrpkg). Those of you willing to have a clean R dependency management should thus have a look at [Packrat](https://rstudio.github.io/packrat/).
Getting the list of installed packages and their version
Finally, it is good to know that there is a built-in R command (`installed.packages`) allowing to retrieve and list the details of all packages installed.
This section is mostly a cut and paste from the [recent post by Ian Pylvainen](https://support.rstudio.com/hc/en-us/articles/219949047-Installing-older-versions-of-packages) on this topic. It comprises a very clear explanation of how to proceed.
### Installing a pre-compiled version
If you're on a Debian or a Ubuntu system, it may be difficult to access a specific version without breaking your system. So unless you are moving to the latest version available in your Linux distribution, **we strongly recommend you to build from source**. In this case, you'll need to make sure you have the necessary toolchain to build packages from source (e.g., gcc, FORTRAN, etc.). On Windows, this may require you to install [Rtools](https://cran.r-project.org/bin/windows/Rtools/).
If you're on Windows or OS X and looking for a package for an **older version of R** (R 2.1 or below), you can check the [CRAN binary archive](https://cran-archive.r-project.org/bin/). Once you have the URL, you can install it using a command similar to the example below:
The simplest method to install the version you need is to use the `install_version()` function of the `devtools` package (obviously, you need to install `devtools` first, which can be done by running in `R` the command `install.packages("devtools")`). For instance:
Alternatively, you may want to install an older package from source If devtools fails or if you do not want to depend on it, you can install it from source via `install.packages()` directed using the right URL. This URL can be obtained by browsing the [CRAN Package Archive](https://cran.r-project.org/src/contrib/Archive).
Once you have the URL, you can install it using a command similar to the example below:
There are a few potential issues that may arise with installing older versions of packages:
- You may be losing functionality or bug fixes that are only present in the newer versions of the packages.
- The older package version needed may not be compatible with the version of R you have installed. In this case, you will either need to downgrade R to a compatible version or update your R code to work with a newer version of the package.
-[Obtenir des informations sur votre dépôt Git](#obtenir-des-informations-sur-votre-dépôt-git)
-[Obtenir des informations sur les librairies Python 3](#obtenir-des-informations-sur-les-librairies-python-3)
-[Obtenir des informations sur votre système](#obtenir-des-informations-sur-votre-système)
-[Lister les packages installés et leur version](#lister-les-packages-installés-et-leur-version)
-[Lister les packages importés (chargés dans une session Python) et leur version](#lister-les-packages-importés-chargés-dans-une-session-python-et-leur-version)
-[Sauvegarder et restaurer un environnement avec pip](#sauvegarder-et-restaurer-un-environnement-avec-pip)
-[Installer un nouveau package ou une version spécifique](#installer-un-nouveau-package-ou-une-version-spécifique)
-[Obtenir des informations sur les packages R](#obtenir-des-informations-sur-les-packages-r)
-[Lister les packages installés et leur version](#lister-les-packages-installés-et-leur-version-1)
-[Lister les packages importés (chargés dans une session R) et leur version](#lister-les-packages-importés-chargés-dans-une-session-r-et-leur-version)
-[Installer un nouveau package ou une version spécifique](#installer-un-nouveau-package-ou-une-version-spécifique-1)
Obtenir des informations sur votre dépôt Git
============================================
Lorsqu'on prend des notes, il peut être difficile de se rappeler quelle version de code ou de fichier a été utilisée. C'est l'intérêt du contrôle de versions. Voici quelques commandes utiles que nous insérons généralement au début de nos notebooks dans des cellules shell :
``` shell
git log -1
```
``` example
commit 741b0088af5b40588493c23c46d6bab5d0adeb33
Author: Arnaud Legrand <arnaud.legrand@imag.fr>
Date: Tue Sep 4 12:45:43 2018 +0200
Fix a few typos and provide information on jupyter-git plugins.
```
``` shell
git status -u
```
``` example
On branch master
Your branch is ahead of 'origin/master' by 4 commits.
(use "git push" to publish your local commits)
Changes not staged for commit:
(use "git add <file>..." to update what will be committed)
(use "git checkout -- <file>..." to discard changes in working directory)
modified: resources.org
Untracked files:
(use "git add <file>..." to include in what will be committed)
no changes added to commit (use "git add" and/or "git commit -a")
```
*Note : L'option -u indique que Git doit aussi afficher le contenu des répertoires nouvellement créés.*
Nous ajoutons aussi souvent des commandes à la fin des notebooks pour indiquer comment commiter les résultats (ajout des nouveaux fichiers, commit avec un message clair et commande push). Par exemple :
``` shell
git add resources.org;
git commit -m"Completing the section on getting Git information"
git push
```
``` example
[master 514fe2c1 ] Completing the section on getting Git information
To gitlab.inria.fr:learninglab/mooc-rr/mooc-rr-ressources.git
6359f8c..1f8a567 master -> master
```
Évidemment, dans ce cas il faut enregistrer le notebook avant d'exécuter cette cellule, donc le résultat de cette dernière commande (avec le nouveau Git hash) ne sera pas stocké dans la cellule. Ce n'est pas vraiment un problème et c'est le prix à payer pour exécuter Git à partir du notebook lui-même.
Obtenir des informations sur les librairies Python 3
Cette question est discutée sur [StackOverflow](https://stackoverflow.com/questions/20180543/how-to-check-version-of-python-modules). Il est facile d'obtenir la version de tous les packages installés en exécutant `pip` (l'installateur de packages Python) dans une commande shell (la commande peut être `pip` or `pip3` selon la configuration de votre ordinateur).
Voici par exemple comment j'obtiens ces informations sur ma machine :
``` shell
pip3 freeze
```
``` example
asn1crypto==0.24.0
attrs==17.4.0
bcrypt==3.1.4
beautifulsoup4==4.6.0
bleach==2.1.3
...
pandas==0.22.0
pandocfilters==1.4.2
paramiko==2.4.0
patsy==0.5.0
pexpect==4.2.1
...
traitlets==4.3.2
tzlocal==1.5.1
urllib3==1.22
wcwidth==0.1.7
webencodings==0.5
```
Dans un notebook Jupyter, cela peut facilement être fait en utilisant la commande magique `%%sh`. Voici par exemple ce que vous pouvez faire et obtenir avec les notebooks Jupyter que nous avons déployés pour le MOOC (notez qu’ici vous devez simplement utiliser la commande `pip`) :
``` python
%%sh
pipfreeze
```
``` example
alembic==0.9.9
asn1crypto==0.24.0
attrs==18.1.0
Automat==0.0.0
...
numpy==1.13.3
olefile==0.45.1
packaging==17.1
pamela==0.3.0
pandas==0.22.0
...
webencodings==0.5
widgetsnbextension==3.2.1
xlrd==1.1.0
zope.interface==4.5.0
```
Dans la suite de ce document, je supposerai que la commande correcte est `pip` et je ne vais pas systématiquement insérer la commande magique `%%sh`.
Une fois que vous savez quels packages sont installés, vous pouvez facilement obtenir des informations supplémentaires sur un package donné et notamment vérifier s'il a été installé "localement" via pip ou s'il est installé à l'échelle du système. À nouveau dans une commande shell :
``` shell
pip show pandas
echo" "
pip show statsmodels
```
``` example
Name: pandas
Version: 0.22.0
Summary: Powerful data structures for data analysis, time series,and statistics
Home-page: http://pandas.pydata.org
Author: None
Author-email: None
License: BSD
Location: /usr/lib/python3/dist-packages
Requires:
Name: statsmodels
Version: 0.9.0
Summary: Statistical computations and models for Python
Vous pouvez vouloir savoir quels packages sont chargés dans une session Python ainsi que leur version sans recourir à pip (qui listera tous les packages disponibles). Inspiré par [StackOverflow](https://stackoverflow.com/questions/4858100/how-to-list-imported-modules), voici une fonction simple qui liste les packages chargés (ayant un attribut `__version__`, qui n’est malheureusement pas tout à fait standard).
``` python
defprint_imported_modules():
importsys
forname,valinsorted(sys.modules.items()):
if(hasattr(val,'__version__')):
print(val.__name__,val.__version__)
else:
print(val.__name__,"(unknown version)")
print("**** Package list in the beginning ****");
print_imported_modules()
print("**** Package list after loading pandas ****");
importpandas
print_imported_modules()
```
``` example
**** Package list in the beginning ****
**** Package list after loading pandas ****
_csv 1.0
_ctypes 1.1.0
decimal 1.70
argparse 1.1
csv 1.0
ctypes 1.1.0
cycler 0.10.0
dateutil 2.7.3
decimal 1.70
distutils 3.6.5rc1
ipaddress 1.0
json 2.0.9
logging 0.5.1.2
matplotlib 2.1.1
numpy 1.14.5
numpy.core 1.14.5
numpy.core.multiarray 3.1
numpy.core.umath b'0.4.0'
numpy.lib 1.14.5
numpy.linalg._umath_linalg b'0.1.5'
pandas 0.22.0
_libjson 1.33
platform 1.0.8
pyparsing 2.2.0
pytz 2018.5
re 2.2.1
six 1.11.0
urllib.request 3.6
zlib 1.0
```
Sauvegarder et restaurer un environnement avec pip
La façon la plus simple de faire est la suivante :
``` shell
pip3 freeze > requirements.txt # to obtain the list of packages with their version
pip3 install-r requirements.txt # to install the previous list of packages, possibly on an other machine
```
Si vous voulez avoir plusieurs environnements Python sur votre ordinateur, vous pouvez vouloir utiliser [Pipenv](https://docs.pipenv.org/)(mais je doute que cela permette de tracer correctement les bibliothèques dynamiques FORTRAN ou C encapsulées dans Python).
Installer un nouveau package ou une version spécifique
L’environnement Jupyter que nous avons déployé sur nos serveurs pour le MOOC est basé sur Miniconda 4.5.4 et Python 3.6. Dans cet environnement, vous devez simplement utiliser la commande `pip` (vous devrez peut-être utiliser `pip3` sur votre ordinateur).
Voici ce que j'obtiens si j'interroge la version actuelle de `statsmodels` dans une commande shell.
``` shell
pip show statsmodels
```
``` example
Name: statsmodels
Version: 0.8.0
Summary: Statistical computations and models for Python
Home-page: http://www.statsmodels.org/
Author: Skipper Seabold, Josef Perktold
Author-email: pystatsmodels@googlegroups.com
License: BSD License
Location: /opt/conda/lib/python3.6/site-packages
Requires: scipy, patsy, pandas
```
Je peux alors facilement mettre `statsmodels` à jour :
``` shell
pip install--upgrade statsmodels
```
La nouvelle version devrait alors être :
``` shell
pip show statsmodels
```
``` example
Name: statsmodels
Version: 0.9.0
Summary: Statistical computations and models for Python
Home-page: http://www.statsmodels.org/
Author: Skipper Seabold, Josef Perktold
Author-email: pystatsmodels@googlegroups.com
License: BSD License
Location: /opt/conda/lib/python3.6/site-packages
Requires: scipy, patsy, pandas
```
Il est même possible d’installer une version spécifique (peut-être beaucoup plus ancienne), par exemple :
``` shell
pip install statsmodels==0.6.1
```
Obtenir des informations sur les packages R
===========================================
Lister les packages installés et leur version
---------------------------------------------
Il existe une commande R intégrée (`installed.packages`) permettant de récupérer et de lister les détails de tous packages installés.
Le meilleur moyen semble être d'utiliser le package `devtools`. Si le package n'est pas installé, vous devez d'abord l'installer en exécutant la commande `install.packages("devtools",dep=TRUE)` dans `R`).
Certains suggèrent même [l'écriture d'un package R pour faciliter la recherche reproductible](https://github.com/ropensci/rrrpkg).
Ceux d'entre vous qui souhaitent disposer d'une gestion des dépendances "propre" en R peuvent aussi jeter un coup d'œil à [Packrat](https://rstudio.github.io/packrat/).
Installer un nouveau package ou une version spécifique
Cette section est principalement un copier-coller du [récent post de Ian Pylvainen](https://support.rstudio.com/hc/en-us/articles/219949047-Installing-older-versions-of-packages) sur ce sujet. Il comprend une explication très claire de la façon de procéder.
### Installer une version pré-compilée
Si vous êtes sur un système Debian ou Ubuntu, il peut être difficile d’accéder à une version spécifique sans casser votre système. Donc, sauf si vous passez à la dernière version disponible pour votre distribution Linux, **nous vous recommandons fortement d'installer les packages à l'aide des fichiers source**. Dans ce cas, vous devez vous assurer que vous disposez de l'ensemble des outils nécessaires pour créer des packages à partir des sources (par exemple : gcc, FORTRAN, etc.). Sous Windows, il se peut que vous deviez installer [Rtools](https://cran.r-project.org/bin/windows/Rtools/).
Si vous utilisez Windows ou OS X et recherchez un package pour une **ancienne version de R** (version 2.1 ou inférieure), vous pouvez le rechercher sur l'[archive binaire du site CRAN](https://cran-archive.r-project.org/bin/). Une fois que vous avez l'URL, vous pouvez l'installer à l'aide d'une commande similaire à l'exemple ci-dessous :
La méthode la plus simple pour installer la version dont vous avez besoin consiste à utiliser la fonction `install_version()` du package `devtools` (vous devez évidemment avoir préalablement installé `devtools`, ce qui peut être fait en exécutant `R` la commande `install.packages("devtools",dep=TRUE)`). Par exemple:
Vous pouvez également installer un ancien package à partir de son code source. Si vous ne réussissez pas avec devtools ou si vous ne voulez pas dépendre de ce package, vous pouvez l’installer à partir du source à l'aide de la commande `install.packages()` en utilisant la bonne URL. Cette URL peut être obtenue en naviguant dans [l'archive de packages du site CRAN](https://cran.r-project.org/src/contrib/Archive).
Une fois que vous avez l'URL, vous pouvez l'installer à l'aide d'une commande similaire à l'exemple ci-dessous :
Quelques problèmes sont susceptibles de survenir lors de l'installation d'anciennes versions de packages :
- Vous pouvez perdre des fonctionnalités ou des corrections de bugs qui ne sont présentes que dans les versions les plus récentes des packages.
- L'ancienne version de package requise peut ne pas être compatible avec la version de R que vous avez installée. Dans ce cas, vous devrez soit rétrograder R vers une version compatible, soit mettre à jour votre code R pour qu'il fonctionne avec une version plus récente du package.
-["Thoughts" on language/software stability](#thoughts-on-languagesoftware-stability)
-[Controlling your software environment](#controlling-your-software-environment)
-[Preservation/Archiving](#preservationarchiving)
-[Workflows](#workflows)
-[Numerical and statistical issues](#numerical-and-statistical-issues)
-[Publication practices](#publication-practices)
-[Experimentation](#experimentation)
"Thoughts" on language/software stability
=========================================
As we explained, the programming language used in an analysis has a clear influence on the reproducibility of your analysis. It is not a characteristic of the language itself but rather a consequence of the development philosophy of the underlying community. For example C is a very stable language with a [very clear specification designed by a committee](https://en.wikipedia.org/wiki/C_(programming_language)#ANSI_C_and_ISO_C) (even though some compilers may not respect this norm).
On the other end of the spectrum, [Python](https://en.wikipedia.org/wiki/Python_(programming_language)) had a much more organic development based on a readability philosophy and valuing continuous improvement over backwards-compatibility. Furthermore, Python is commonly used as a wrapping language (e.g., to easily use C or FORTRAN libraries) and has its own packaging system. All these design choices tend to make reproducibility often a bit painful with Python, even though the community is slowly taking this into account. The transition from Python 2 to the not fully backwards compatible Python 3 has been a particularly painful process, not least because the two languages are so similar that is it not always easy to figure out if a given script or module is written in Python 2 or Python 3. It isn't even rare to see Python scripts that work under both Python 2 and Python 3, but produce different results due to the change in the behavior of integer division.
[R](https://en.wikipedia.org/wiki/R_(programming_language)), in comparison is much closer (in terms of developer community) to languages like [SAS](https://en.wikipedia.org/wiki/SAS_(software)), which is heavily used in the pharmaceutical industry where statistical procedures need to be standardized and rock solid/stable. R is obviously not immune to evolutions that break old versions and hinder reproducibility/backward compatibility. Here is a relatively recent [true story about this](http://members.cbio.mines-paristech.fr/~thocking/HOCKING-reproducible-research-with-R.html) and some colleagues who worked on the [statistics introductory course with R on FUN](https://www.fun-mooc.fr/courses/UPSUD/42001S06/session06/about) reported us several issues with a few functions (`plotmeans` from `gplots`, `survfit` from `survival`, or `hclust`) whose default parameters had changed over the years. It is thus probably good practice to give explicit values for all parameters (which can be cumbersome) instead of relying on default values, and to restrict your dependencies as much as possible.
This being said, the R development community is generally quite careful about stability. We (the authors of this MOOC) believe that open source (which allows to inspect how computation is done and to identify both mistakes and sources of non-reproducibility) is more important than the rock solid stability of SAS, which is proprietary software.
Yet, if you really need to stay with SAS, you should know that SAS can be used within Jupyter using the [Python SASPy](https://sassoftware.github.io/saspy/) and the [Python SASKernel](https://sassoftware.github.io/sas_kernel/) packages (step by step explanations about this are given [here](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/documents/tuto_jupyter_windows/tuto_jupyter_windows.md#53-le-package-python-saspy-permet-dex%C3%A9cuter-du-code-sas-dans-un-notebook-python)). Using such literate programming approach allied with systematic version and environment control will always help. Similar solutions exist for many languages ([list of Jupyter kernels](https://github.com/jupyter/jupyter/wiki/Jupyter-kernels)).
Controlling your software environment
=====================================
As we mentioned in the video sequences, there are several solutions to control your environment:
- The easy (preserve the mess) ones: [CDE](http://www.pgbovine.net/cde.html) or [ReproZip](https://vida-nyu.github.io/reprozip/)
- The more demanding (encourage cleanliness) where you start with a clean environment and install only what's strictly necessary (and document it):
- The very well known [Docker](https://www.docker.io/)
-[Singularity](https://singularity.lbl.gov/) or [Spack](https://spack.io/), which are more targeted toward the specific needs of high performance computing users
-[Guix](https://www.gnu.org/software/guix/), [Nix](https://nixos.org/) that are very clean (perfect?) solutions to this dependency hell and which we recommend
It may be hard to understand the difference between these different approaches and decide which one is better in your context.
Here is a webinar where some of these tools are demoed in a reproducible research context: [Controling your environment (by Michael Mercier and Cristian Ruiz)](https://github.com/alegrand/RR_webinars/blob/master/2_controling_your_environment/index.org)
You may also want to have a look at [the Popper conventions](http://falsifiable.us/)([webinar by Ivo Gimenez through google hangout](https://github.com/alegrand/RR_webinars/blob/master/11_popper/index.org)) or at the [presentation of Konrad Hinsen on Active Papers](https://github.com/alegrand/RR_webinars/blob/master/7_publications/index.org)(<http://www.activepapers.org/>).
Preservation/Archiving
======================
Ensuring software is properly archived, i.e, is safely stored so that it can be accessed in a perennial way, can be quite tricky. If you have never seen [Roberto Di Cosmo presenting the Software Heritage project](https://github.com/alegrand/RR_webinars/blob/master/5_archiving_software_and_data/index.org), this is a must see. [<https://www.softwareheritage.org/>](https://www.softwareheritage.org/)
For regular data, we highly recommend using [<https://www.zenodo.org/>](https://www.zenodo.org/) whenever the data is not sensitive.
Workflows
=========
In the video sequences, we mentioned workflow managers (original application domain in parenthesis):
- Light-weight: [dask](http://dask.pydata.org/)(python), [drake](https://ropensci.github.io/drake/)(R), [swift](http://swift-lang.org/)(molecular biology), [snakemake](https://snakemake.readthedocs.io/)(like`make` but more expressive and in `python`)...
You may want to have a look at this webinar: \[\[<https://github.com/alegrand/RR_webinars/blob/master/6_reproducibility_bioinformatics/index.org>\]\[Reproducible Science in Bio-informatics: Current Status, Solutions and Research Opportunities (by Sarah Cohen Boulakia, Yvan Le Bras and Jérôme Chopard).\]\]
Numerical and statistical issues
================================
We have mentioned these topics in our MOOC but we could by no way cover them properly. We only suggest here a few interesting talks about this.
-\[\[<https://github.com/alegrand/RR_webinars/blob/master/10_statistics_and_replication_in_HCI/index.org>\]\[In this talk, Pierre Dragicevic provides a nice illustration of the consequences of statistical uncertainty and of how some concepts (e.G. p-values) are commonly badly understood.\]\]
-\[\[<https://github.com/alegrand/RR_webinars/blob/master/3_numerical_reproducibility/index.org>\]\[Nathalie Revol, Philippe Langlois and Stef Graillat present the main challenges encountered when trying to achieve numerical reproducibility and present recent research work on this topic.\]\]
Publication practices
=====================
You may want to have a look at the following two webinars:
-[Enabling open and reproducible research at computer systems’ conferences (by Grigori Fursin)](https://github.com/alegrand/RR_webinars/blob/master/8_artifact_evaluation/index.org). In particular, this talk discusses *artifact evaluation* that is becoming more and more popular.
-[Publication Modes Favoring Reproducible Research (by Konrad Hinsen and Nicolas Rougier)](https://github.com/alegrand/RR_webinars/blob/master/7_publications/index.org). In this talk, the motivation for the [ReScience journal](http://rescience.github.io/) initiative are presented.
-[Simine Vazire - When Should We be Skeptical of Scientific Claims?](https://www.youtube.com/watch?v=HuJ2G8rXHMs), which is discussing publication practices in social sciences and in particular HARKing (Hypothesizing After the Results are Known), p-hacking, etc.
Experimentation
===============
Experimentation was not covered in this MOOC, although it is an essential part of science. The main reason is that practices and constraints can vary so wildly from one domain to another that it could not be properly covered in a first edition. We would be happy to gather references you consider as interesting in your domain so do not hesitate to provide us with such references by using the forum and we will update this page.
-[A recent talk by Lucas Nussbaum on Experimental Testbeds in Computer Science](https://github.com/alegrand/RR_webinars/blob/master/9_experimental_testbeds/index.org).
Comme nous l'avons expliqué, le langage de programmation utilisé dans une analyse a une influence évidente sur la reproductibilité de l'analyse. Ce n'est pas une caractéristique du langage lui-même mais plutôt une conséquence de la philosophie de développement de la communauté sous-jacente. Par exemple, C est un langage très stable avec une [spécification très claire conçue par un comité](https://en.wikipedia.org/wiki/C_(programming_language)#ANSI_C_and_ISO_C) (même si certains compilateurs ne respectent cette norme).
À l’autre bout du spectre, [Python](https://en.wikipedia.org/wiki/Python_(programming_language)) a connu un développement beaucoup plus organique basé sur une philosophie de lisibilité et valorisant l'amélioration continue par rapport à la compatibilité ascendante. De plus, Python est couramment utilisé comme langage de wrapping (par exemple pour utiliser facilement les bibliothèques C ou FORTRAN) et dispose de son propre système de packaging. Tous ces choix de conception ont souvent tendance à rendre la reproductibilité un peu pénible avec Python, même si la communauté en tient lentement compte. La transition de Python 2 à Python 3, qui n'est pas totalement compatible avec les versions antérieures, a été un processus particulièrement pénible, notamment parce que les deux langages sont si similaires qu'il n'est pas toujours facile de savoir si un script ou un module donné est écrit en Python 2 ou Python 3. Il n'est même pas rare de voir des scripts Python qui fonctionnent sous Python 2 et Python 3, mais produisent des résultats différents en raison du changement de comportement de la division entière.
[R](https://en.wikipedia.org/wiki/R_(programming_language)), en comparaison, est beaucoup plus proche (en termes de communauté de développeurs) de langages comme [SAS](https://en.wikipedia.org/wiki/SAS_(logiciel)), très utilisé dans l'industrie pharmaceutique où les procédures statistiques doivent être standardisées et stables/solides. R n’est évidemment pas à l’abri des évolutions qui cassent les anciennes versions et gênent la reproductibilité/compatibilité avec les versions antérieures. Voici une [véritable histoire relativement récente à ce sujet](http://members.cbio.mines-paristech.fr//thocking/HOCKING-reproducible-research-with-R.html) et des collègues qui ont travaillé sur le [MOOC d'initiation à la statistique avec R sur FUN](https://www.fun-mooc.fr/courses/course-v1:UPSUD+42001+session10/about) nous ont signalé plusieurs problèmes concernant quelques fonctions (`gplots::plotmeans`, `survival::survfit` ou `hclust`) dont les paramètres par défaut ont changé au fil des ans. Il est donc probablement utile de donner des valeurs explicites pour tous les paramètres (ce qui peut être fastidieux) au lieu de s’appuyer sur des valeurs par défaut et de restreindre autant que possible vos dépendances.
Ceci étant dit, la communauté de développement R est généralement très prudente en matière de stabilité. Nous (les auteurs de ce MOOC) pensons que l'open source (qui permet de contrôler le calcul et d'identifier les erreurs et les sources de non-reproductibilité) est plus important que la stabilité à toute épreuve de SAS, qui est un logiciel propriétaire.
Cependant, si vous avez vraiment besoin de rester sous SAS, sachez que SAS peut être utilisé dans Jupyter en utilisant les packages Python [SASPy](https://sassoftware.github.io/saspy/) et [SASKernel](https://sassoftware.github.io/sas_kernel/)(tuto pas à pas [ici](https://gitlab.inria.fr/learninglab/mooc-rr/mooc-rr-ressources/blob/master/documents/tuto_jupyter_windows/tuto_jupyter_windows.md#53-le-package-python-saspy-permet-dex%C3%A9cuter-du-code-sas-dans-un-notebook-python)). L'utilisation d'une telle approche de programmation alphabète alliée à un contrôle systématique de la version et de l'environnement est un plus. Des solutions similaires existent pour de nombreux langages ([liste des kernels Jupyter](https://github.com/jupyter/jupyter/wiki/Jupyter-kernels)).
Contrôler votre environnement logiciel
======================================
Comme nous l'avons mentionné dans les séquences vidéo, plusieurs solutions permettent de contrôler votre environnement :
- Les faciles (autorisant le désordre) : [CDE](http://www.pgbovine.net/cde.html) ou [ReproZip](https://vida-nyu.github.io/reprozip/)
- Les plus exigeants (encourageant la propreté) où vous commencez avec un environnement propre et n’installez que ce qui est strictement nécessaire (et le documentez) :
- Le très connu [Docker](https://www.docker.io/)
-[Singularity](https://singularity.lbl.gov/) ou [Spack](https://spack.io/), qui sont plus ciblés sur les besoins spécifiques des utilisateurs d'informatique de haute performance
-[Guix](https://www.gnu.org/software/guix/), [Nix](https://nixos.org/) qui sont des solutions très propres (parfaites ?) à cet enfer de dépendance et que nous recommandons
Il peut être difficile de comprendre la différence entre ces différentes approches et de décider laquelle est la meilleure dans votre contexte.
Voici un webinaire où certains de ces outils sont présentés dans un contexte de recherche reproductible : [Controling your environment (by Michael Mercier and Cristian Ruiz)](https://github.com/alegrand/RR_webinars/blob/master/2_controling_your_environment/index.org).
Vous voudrez peut-être aussi jeter un coup d’œil sur la convention [Popper](http://falsifiable.us/)([webinaire par Ivo Gimenez par Google hangout](https://github.com/alegrand/RR_webinars/blob/master/11_popper/index.org)) ou sur la [présentation de Konrad Hinsen](https://github.com/alegrand/RR_webinars/blob/master/7_publications/index.org) sur [Active Papers](http://www.activepapers.org/).
Préservation/Archivage
======================
S'assurer que le logiciel est correctement archivé, c'est-à-dire qu'il est stocké de manière sécurisée afin de pouvoir y accéder de manière pérenne, peut être assez délicat. Si vous n'avez jamais vu la [présentation du projet Software Heritage par Roberto Di Cosmo](https://github.com/alegrand/RR_webinars/blob/master/5_archiving_software_and_data/index.org), c'est à voir absolument. [<https://www.softwareheritage.org/>](https://www.softwareheritage.org/)
Pour les données, nous vous recommandons vivement d’utiliser [<https://www.zenodo.org/>](https://www.zenodo.org/) lorsque les données ne sont pas sensibles.
Workflows
=========
Dans les séquences vidéo, nous avons mentionné les gestionnaires de flux de travaux (domaine d'application d'origine entre parenthèses) :
- workflows assez complexes : [Galaxy](https://galaxyproject.org/)(génomique), [Kepler](https://kepler-project.org/)(écologie), [Taverna](https://taverna.apache.org/)(bioinformatique), [Pegasus](https://pegasus.isi.edu/)(astronomie), [Collective Knowledge](http://cknowledge.org/)(optimisation de la compilation), [VisTrails](https://www.vistrails.org)(traitement d'image)
- workflows plus légers : [dask](http://dask.pydata.org/)(Python), [drake](https://ropensci.github.io/drake/)(R), [swift](http://swift-lang.org/)(biologie moléculaire), [snakemake](https://snakemake.readthedocs.io/)(comme`make` mais plus expressif et en `Python`)...
- hybride à mi-chemin entre le notebook et le workflow : [SOS-notebook](https://vatlab.github.io/sos-docs/)
Vous voudrez peut-être consulter ce webinaire : \[\[<https://github.com/alegrand/RR_webinars/blob/master/6_reproducibility_bioinformatics/index.org>\]\[Science Reproducible en Bioinformatique : état actuel, solutions et opportunités de recherche (par Sarah Cohen Boulakia, Yvan Le Bras and Jérôme Chopard)\]\].
Problèmes numériques et statistiques
====================================
Nous avons mentionné ces sujets dans notre MOOC mais nous ne pourrions en aucun cas les couvrir correctement. Nous ne suggérons ici que quelques présentations intéressantes à ce sujet.
-\[\[<https://github.com/alegrand/RR_webinars/blob/master/10_statistics_and_replication_in_HCI/index.org>\]\[Pierre Dragicevic donne une belle illustration des conséquences de l’incertitude statistique et de la manière dont certains concepts (par exemple les p-values) sont généralement mal compris\]\].
-\[\[<https://github.com/alegrand/RR_webinars/blob/master/3_numerical_reproducibility/index.org>\]\[Nathalie Revol, Philippe Langlois et Stef Graillat présentent les principaux problèmes rencontrés en essayant d'obtenir une reproductibilité numérique et présentent les travaux de recherche récents en la matière\]\].
Pratiques de publication
========================
Vous souhaiterez peut-être consulter les webinaires suivants :
-[Permettre une recherche ouverte et reproductible lors de conférences sur les systèmes informatiques (par Grigori Fursin)](https://github.com/alegrand/RR_webinars/blob/master/8_artifact_evaluation/index.org). En particulier, cette discussion aborde *l'évaluation des artefacts* qui devient de plus en plus populaire.
-[Modalités de publication favorisant la recherche reproductible (par Konrad Hinsen et Nicolas Rougier)](https://github.com/alegrand/RR_webinars/blob/master/7_publications/index.org). Présentation des motivations de l'intiative [ReScience journal](http://rescience.github.io/).
-[Simine Vazire - Quand devrions-nous être sceptiques face aux affirmations scientifiques ?](https://www.youtube.com/watch?v=HuJ2G8rXHMs), qui traite des pratiques de publication dans les sciences sociales et en particulier HARKing (Hypothèses après que les résultats sont connus), p-hacking, etc.
Expérimentation
===============
L'expérimentation n'est pas couverte dans ce MOOC bien qu'il s'agisse d'un élément essentiel de la science. La raison principale est que les pratiques et les contraintes varient tellement d'un domaine à l'autre que ce thème ne pouvait pas être correctement couvert dans une première édition. Nous serions heureux de rassembler les références que vous jugez intéressantes dans votre domaine. N'hésitez donc pas à nous les fournir à l'aide du forum. Nous les intégrerons dans cette page.
-[Une récente conférence de Lucas Nussbaum sur les bancs d’essais expérimentaux en informatique](https://github.com/alegrand/RR_webinars/blob/master/9_experimental_testbeds/index.org).